S6 HPTM-transcription dynamics
6.1 H3K27me3
6.1.1 Gene bodies
##
## Call:
## glm(formula = H3K27me3_GB ~ Transcription + Group, family = "binomial",
## data = corrdat, na.action = na.exclude)
##
## Coefficients:
## Estimate Std. Error z value Pr(>|z|)
## (Intercept) 0.23128 0.01836 12.600 <2e-16 ***
## Transcription 1.52362 0.94428 1.614 0.107
## Groupb1W -0.48789 0.02589 -18.843 <2e-16 ***
## Groupb2Q -0.46453 0.02586 -17.960 <2e-16 ***
## Groupb2W -0.53712 0.02593 -20.717 <2e-16 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## (Dispersion parameter for binomial family taken to be 1)
##
## Null deviance: 66993 on 48491 degrees of freedom
## Residual deviance: 66423 on 48487 degrees of freedom
## AIC: 66433
##
## Number of Fisher Scoring iterations: 3## OR 2.5 % 97.5 %
## (Intercept) 1.2602085 1.2157024 1.3064034
## Transcription 4.5887869 0.8272917 37.1403134
## Groupb1W 0.6139213 0.5835273 0.6458649
## Groupb2Q 0.6284311 0.5973523 0.6610940
## Groupb2W 0.5844272 0.5554545 0.6148758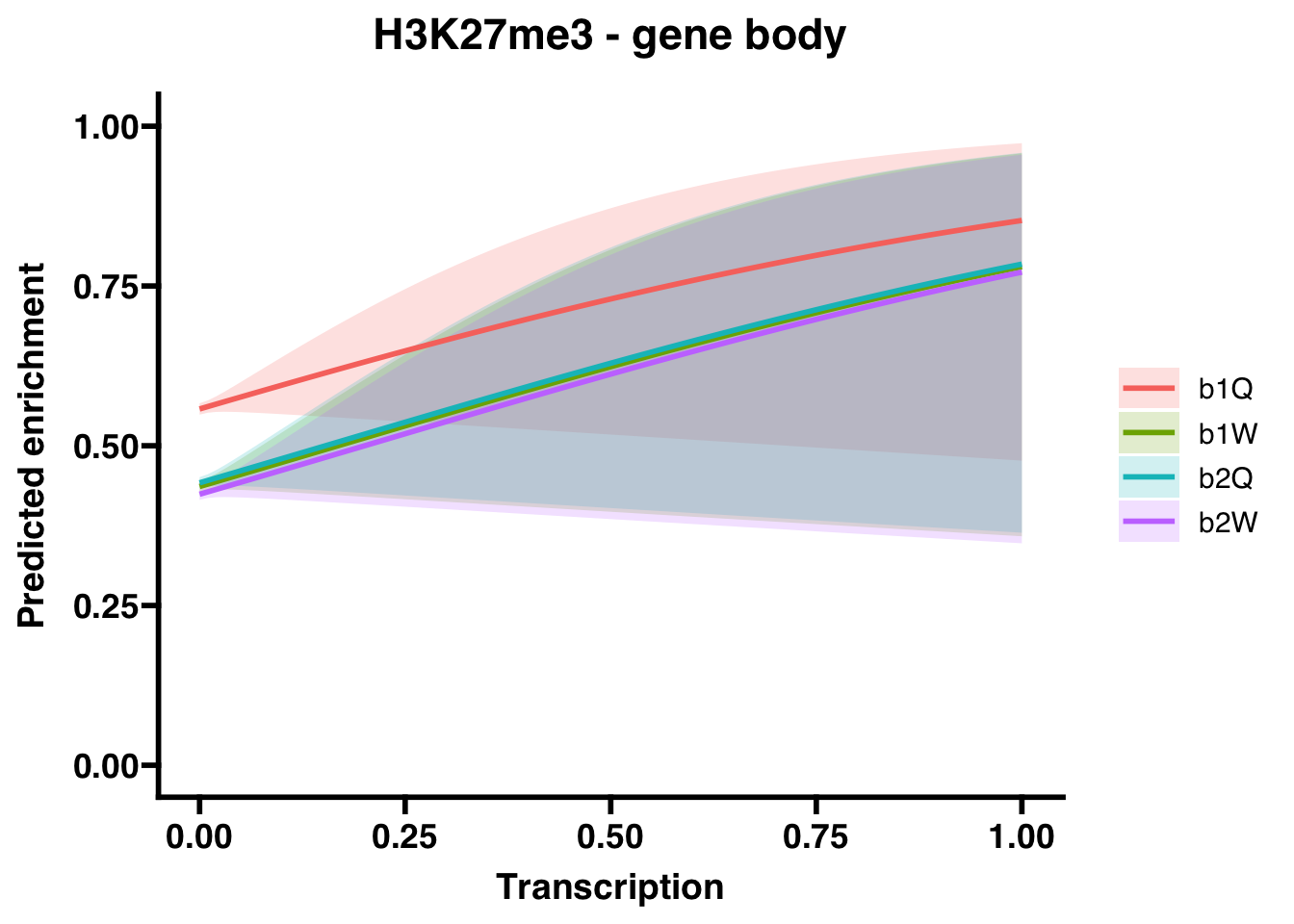
6.1.2 Promoters
##
## Call:
## glm(formula = H3K27me3_P ~ Transcription + Group, family = "binomial",
## data = corrdat, na.action = na.exclude)
##
## Coefficients:
## Estimate Std. Error z value Pr(>|z|)
## (Intercept) -0.19269 0.01842 -10.462 <2e-16 ***
## Transcription -3.50202 1.53037 -2.288 0.0221 *
## Groupb1W -0.43184 0.02643 -16.341 <2e-16 ***
## Groupb2Q -0.37462 0.02629 -14.249 <2e-16 ***
## Groupb2W -0.43267 0.02641 -16.384 <2e-16 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## (Dispersion parameter for binomial family taken to be 1)
##
## Null deviance: 64247 on 48491 degrees of freedom
## Residual deviance: 63863 on 48487 degrees of freedom
## AIC: 63873
##
## Number of Fisher Scoring iterations: 4## OR 2.5 % 97.5 %
## (Intercept) 0.82474140 0.795490181 0.8550473
## Transcription 0.03013634 0.001127849 0.4054232
## Groupb1W 0.64931347 0.616521187 0.6838123
## Groupb2Q 0.68755015 0.653001844 0.7238930
## Groupb2W 0.64877540 0.616032490 0.6832215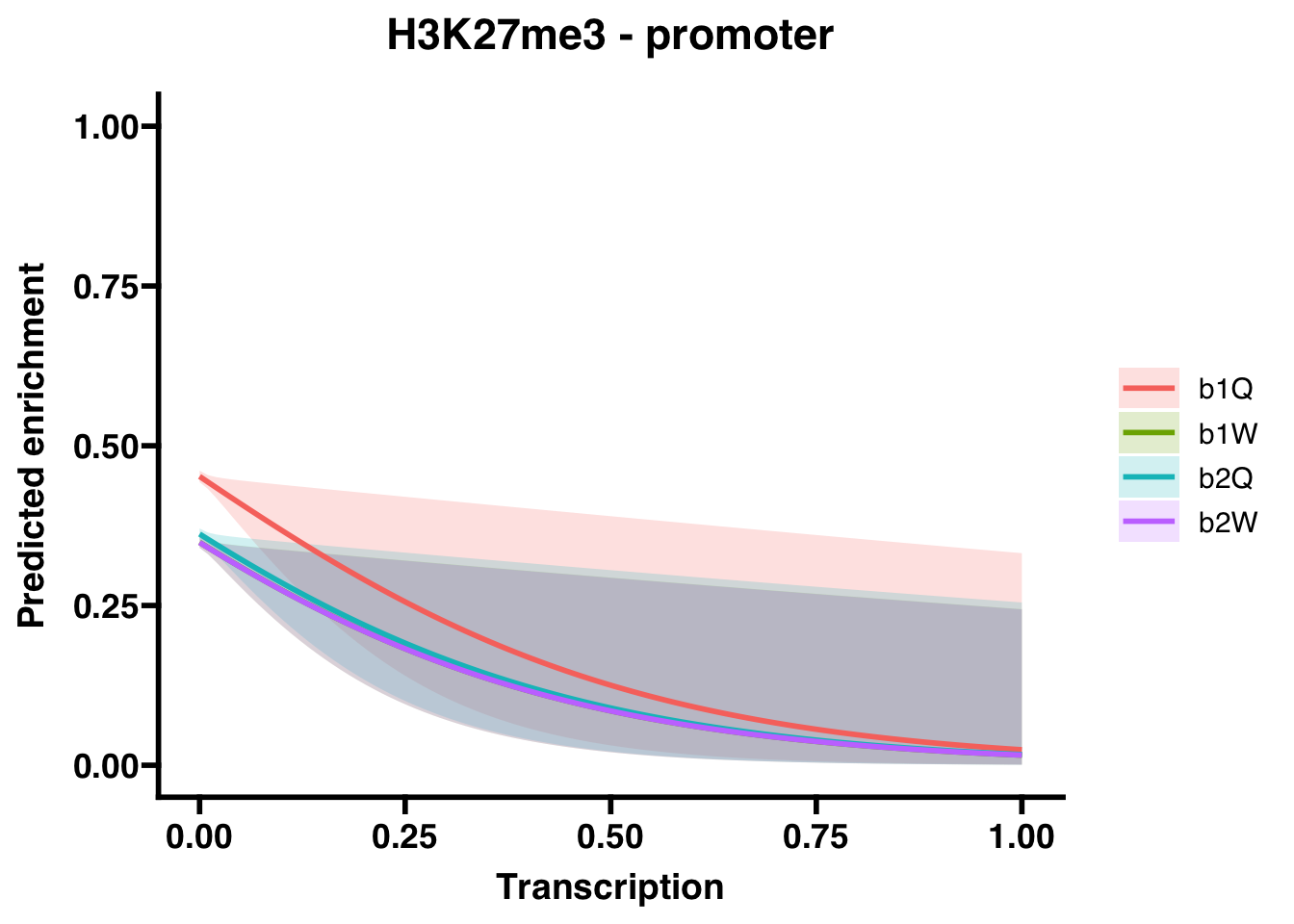
6.2 H3K27ac
6.2.1 Gene bodies
##
## Call:
## glm(formula = H3K27ac_GB ~ Transcription + Group, family = "binomial",
## data = corrdat, na.action = na.exclude)
##
## Coefficients:
## Estimate Std. Error z value Pr(>|z|)
## (Intercept) 0.26665 0.01852 14.400 < 2e-16 ***
## Transcription 5.86657 1.65313 3.549 0.000387 ***
## Groupb1W -0.22906 0.02585 -8.861 < 2e-16 ***
## Groupb2Q -0.42429 0.02586 -16.410 < 2e-16 ***
## Groupb2W -0.73401 0.02616 -28.057 < 2e-16 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## (Dispersion parameter for binomial family taken to be 1)
##
## Null deviance: 67163 on 48491 degrees of freedom
## Residual deviance: 66279 on 48487 degrees of freedom
## AIC: 66289
##
## Number of Fisher Scoring iterations: 4## OR 2.5 % 97.5 %
## (Intercept) 1.3055858 1.2590807 1.353875e+00
## Transcription 353.0366507 17.5064483 1.059278e+04
## Groupb1W 0.7952834 0.7559861 8.366032e-01
## Groupb2Q 0.6542313 0.6218882 6.882252e-01
## Groupb2W 0.4799784 0.4559689 5.052116e-01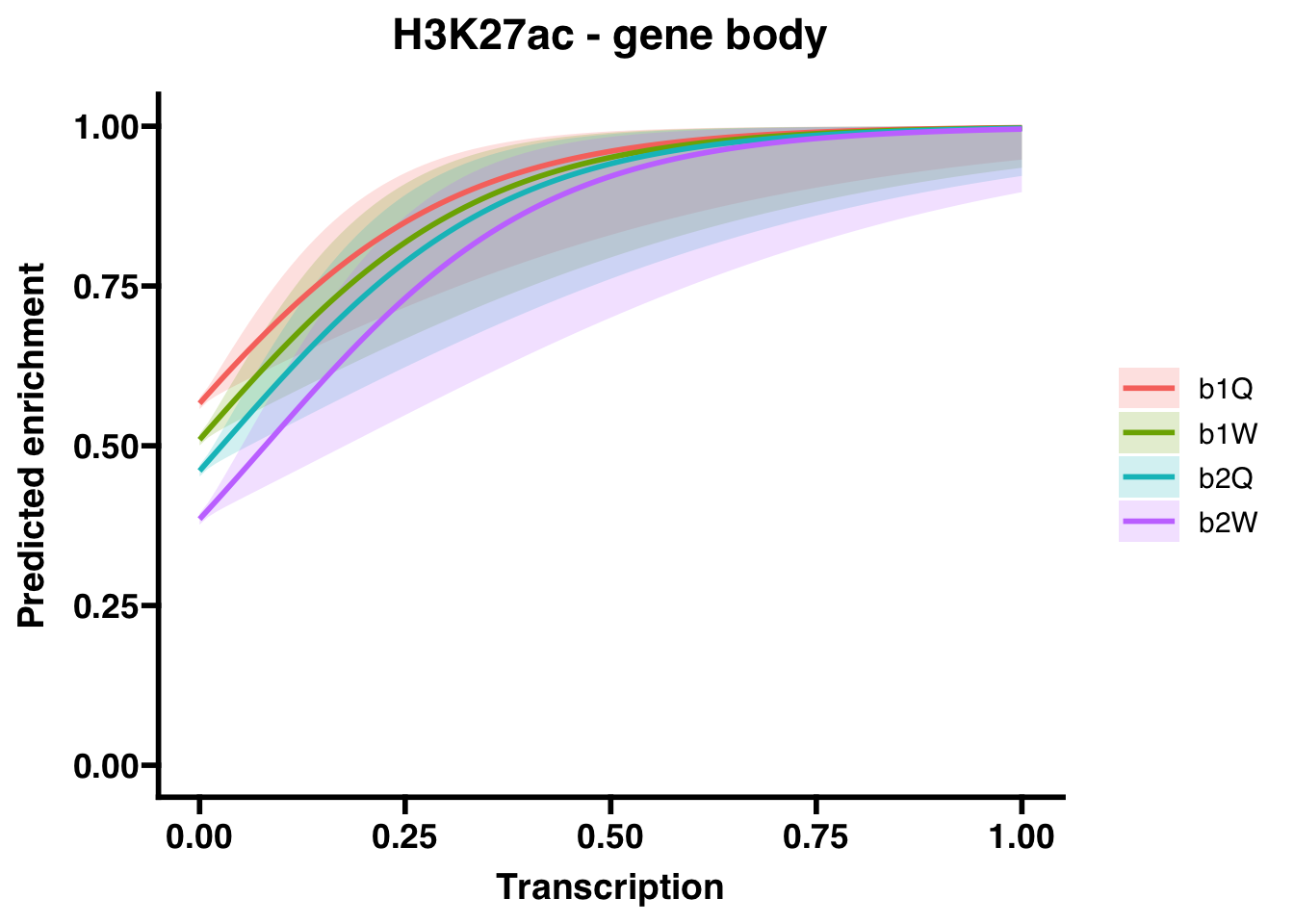
6.2.2 Promoters
##
## Call:
## glm(formula = H3K27ac_P ~ Transcription + Group, family = "binomial",
## data = corrdat, na.action = na.exclude)
##
## Coefficients:
## Estimate Std. Error z value Pr(>|z|)
## (Intercept) -0.12084 0.01825 -6.620 3.58e-11 ***
## Transcription 0.63709 0.83078 0.767 0.44317
## Groupb1W -0.08051 0.02579 -3.122 0.00179 **
## Groupb2Q -0.32665 0.02604 -12.546 < 2e-16 ***
## Groupb2W -0.58884 0.02654 -22.187 < 2e-16 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## (Dispersion parameter for binomial family taken to be 1)
##
## Null deviance: 65645 on 48491 degrees of freedom
## Residual deviance: 65037 on 48487 degrees of freedom
## AIC: 65047
##
## Number of Fisher Scoring iterations: 4## OR 2.5 % 97.5 %
## (Intercept) 0.8861782 0.8550249 0.9184428
## Transcription 1.8909677 0.3623550 10.5315474
## Groupb1W 0.9226433 0.8771682 0.9704675
## Groupb2Q 0.7213358 0.6854358 0.7590876
## Groupb2W 0.5549721 0.5268221 0.5845828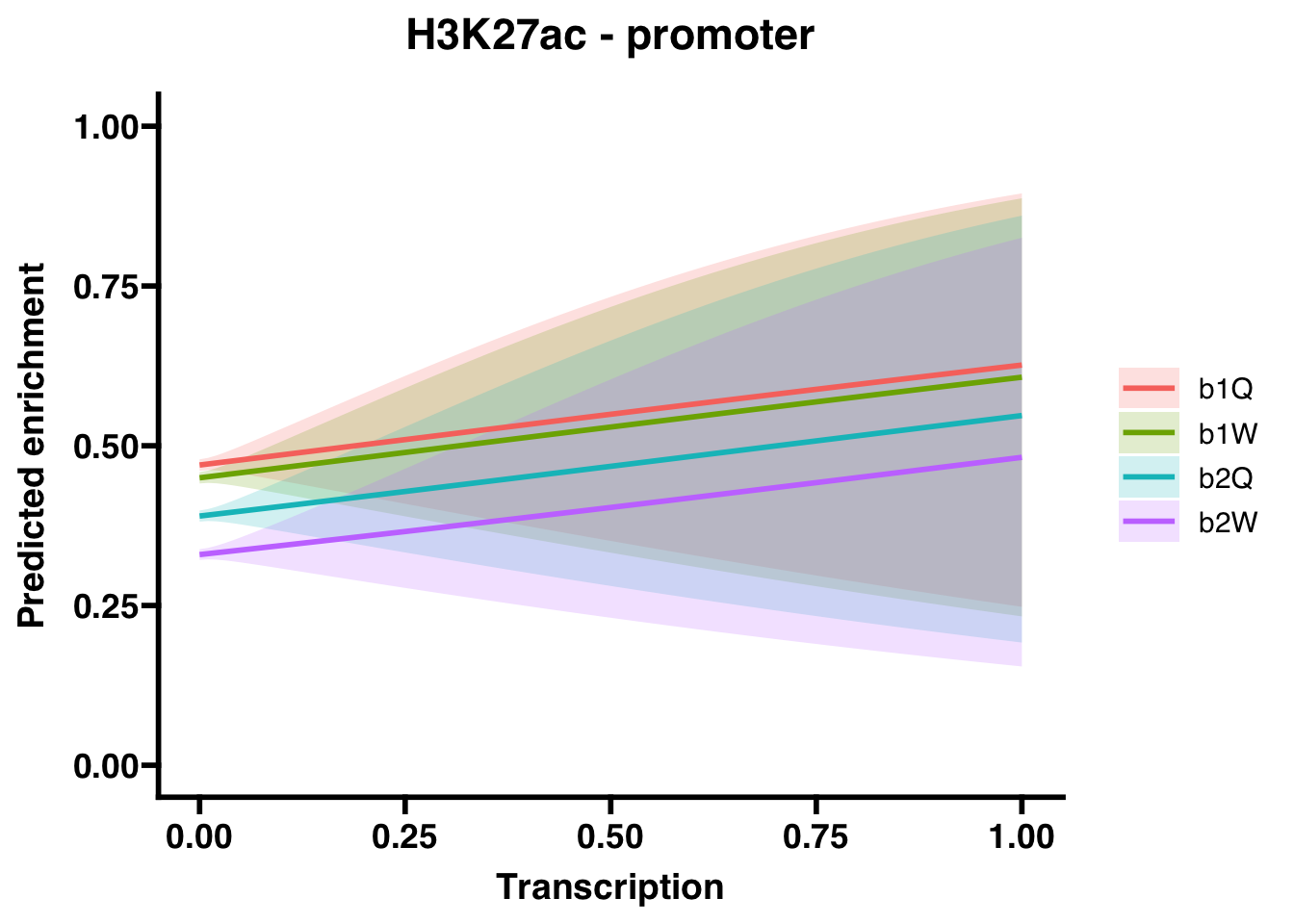
6.3 H3K4me3
6.3.1 Gene bodies
##
## Call:
## glm(formula = H3K4me3_GB ~ Transcription + Group, family = "binomial",
## data = corrdat, na.action = na.exclude)
##
## Coefficients:
## Estimate Std. Error z value Pr(>|z|)
## (Intercept) 0.34155 0.01865 18.310 < 2e-16 ***
## Transcription 6.89945 1.80374 3.825 0.000131 ***
## Groupb1W -0.06428 0.02606 -2.467 0.013635 *
## Groupb2Q -0.31404 0.02590 -12.124 < 2e-16 ***
## Groupb2W -0.27871 0.02591 -10.758 < 2e-16 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## (Dispersion parameter for binomial family taken to be 1)
##
## Null deviance: 66811 on 48491 degrees of freedom
## Residual deviance: 66571 on 48487 degrees of freedom
## AIC: 66581
##
## Number of Fisher Scoring iterations: 4## OR 2.5 % 97.5 %
## (Intercept) 1.4071207 1.3566493 1.459566e+00
## Transcription 991.7309448 35.1738630 3.969657e+04
## Groupb1W 0.9377463 0.8910530 9.868794e-01
## Groupb2Q 0.7304884 0.6943170 7.685177e-01
## Groupb2W 0.7567566 0.7192762 7.961654e-01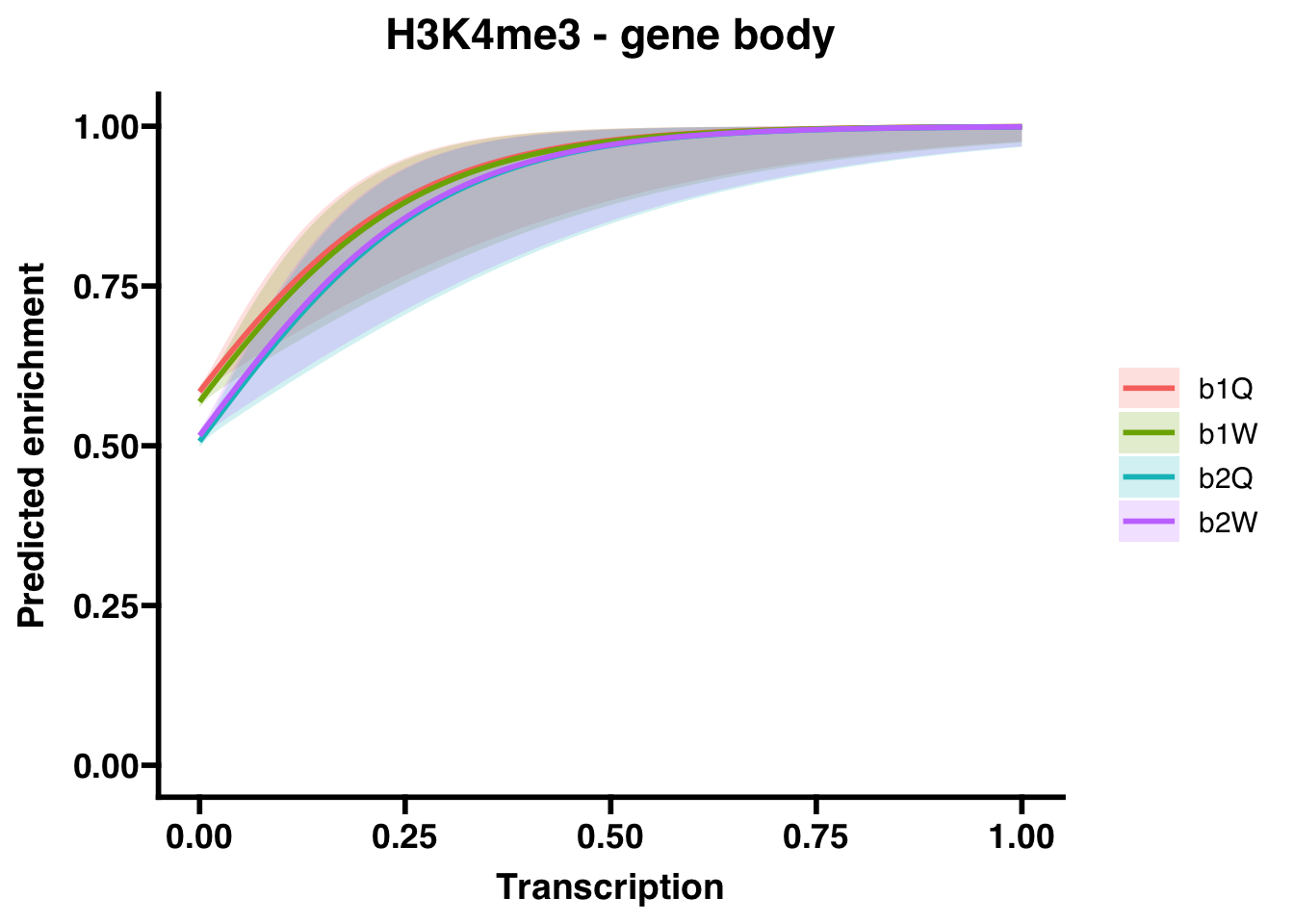
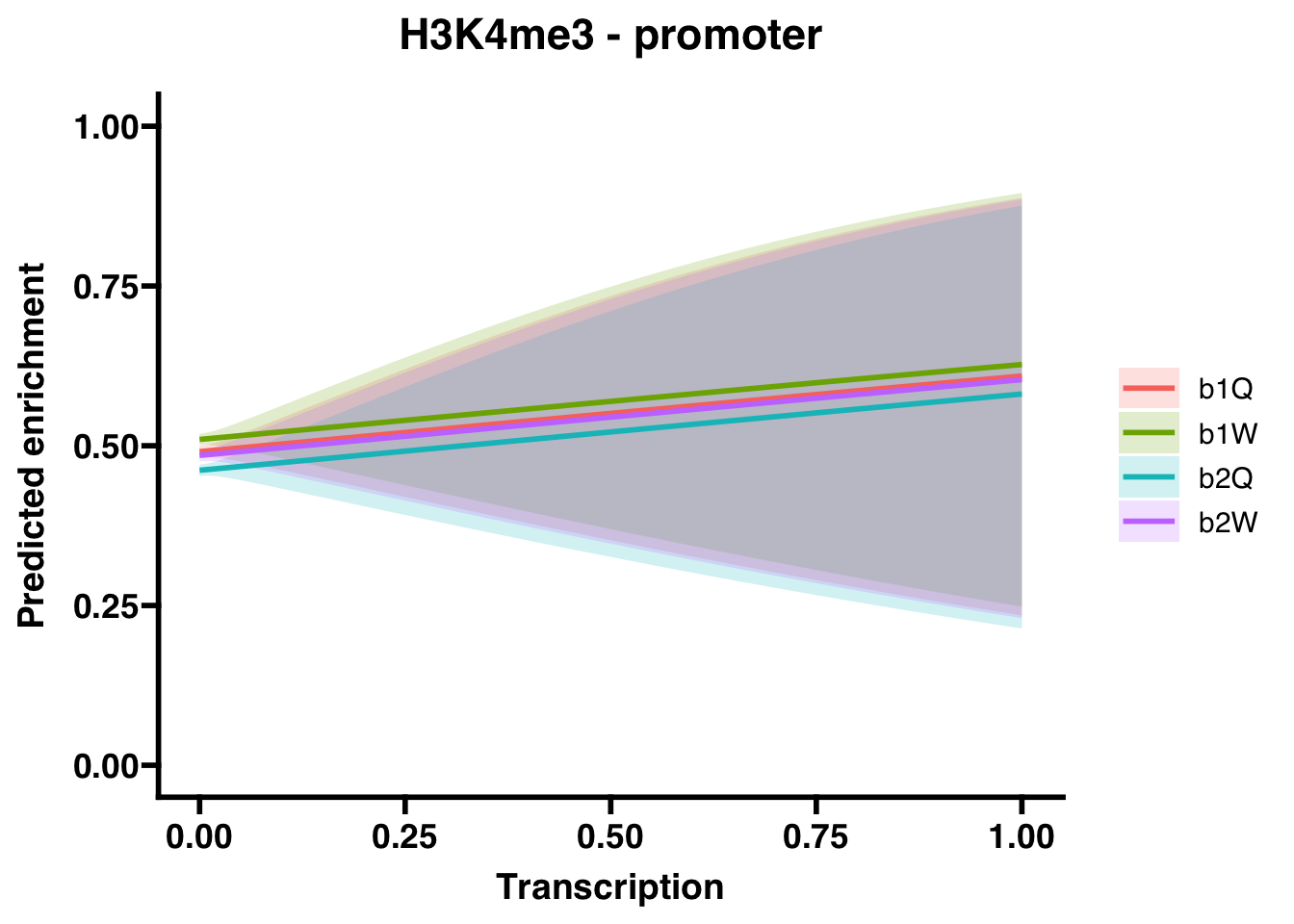
6.3.2 Promoters
##
## Call:
## glm(formula = H3K4me3_P ~ Transcription + Group, family = "binomial",
## data = corrdat, na.action = na.exclude)
##
## Coefficients:
## Estimate Std. Error z value Pr(>|z|)
## (Intercept) -0.03695 0.01822 -2.028 0.04257 *
## Transcription 0.48051 0.83121 0.578 0.56321
## Groupb1W 0.07629 0.02570 2.968 0.00299 **
## Groupb2Q -0.11694 0.02573 -4.545 5.5e-06 ***
## Groupb2W -0.02362 0.02570 -0.919 0.35800
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## (Dispersion parameter for binomial family taken to be 1)
##
## Null deviance: 67191 on 48491 degrees of freedom
## Residual deviance: 67133 on 48487 degrees of freedom
## AIC: 67143
##
## Number of Fisher Scoring iterations: 3## OR 2.5 % 97.5 %
## (Intercept) 0.9637213 0.9299038 0.9987600
## Transcription 1.6168956 0.3181529 9.2732218
## Groupb1W 1.0792802 1.0262608 1.1350481
## Groupb2Q 0.8896392 0.8458815 0.9356488
## Groupb2W 0.9766560 0.9286814 1.0271063