Appendix D-7 POE HPTM enrichment-POE transcription dynamics
7.1 Block 1
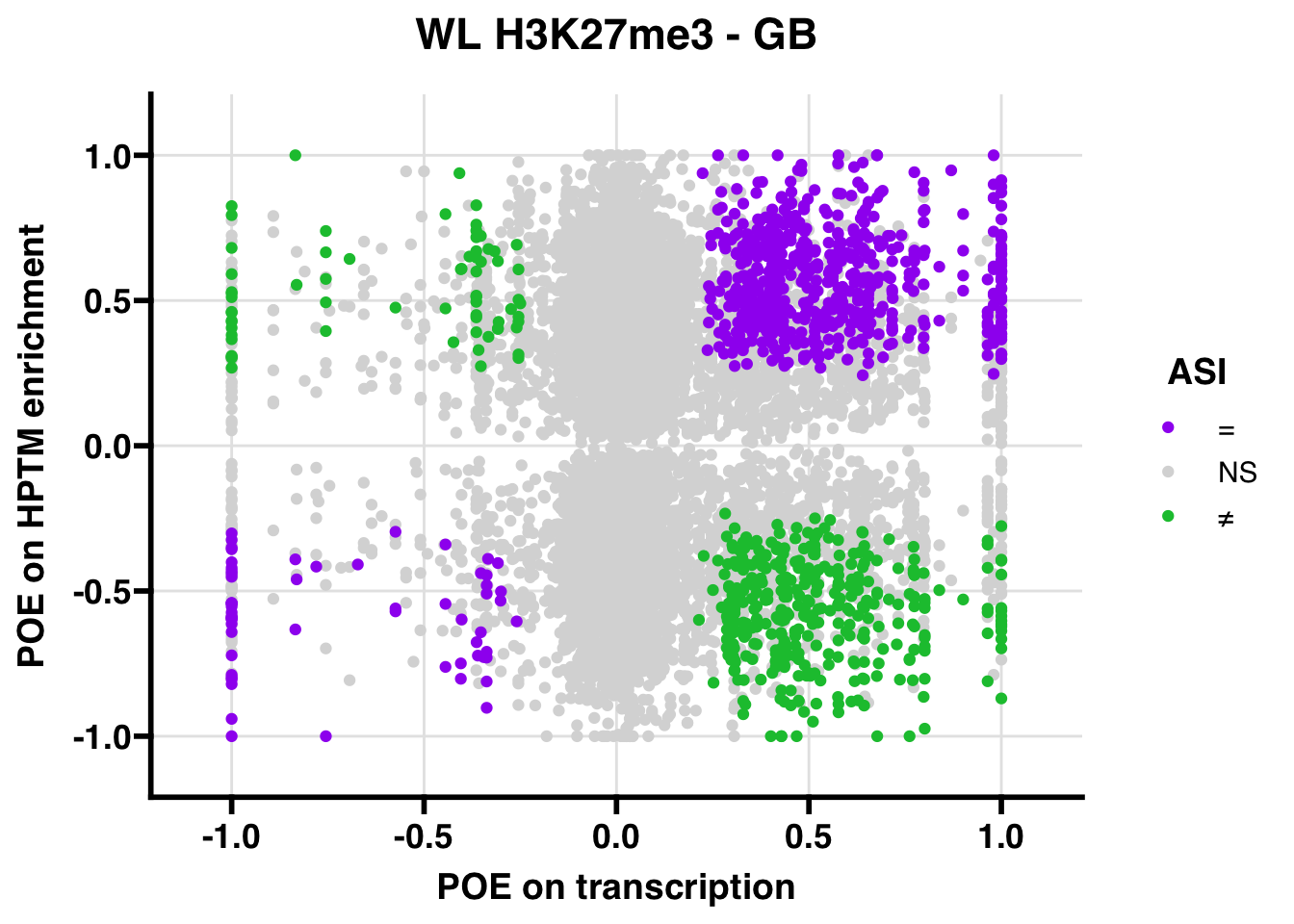
H3K27me3
Gene body
WL
Gene:Peak (categories of allelic bias)
##
## Lineage A:Lineage A Lineage A:NA Lineage A:pat Lineage B:Lineage A
## 1 4 1 11
## Lineage B:Lineage B Lineage B:mat Lineage B:NA Lineage B:pat
## 2 7 40 5
## mat:Lineage A mat:Lineage B mat:mat mat:NA
## 7 1 9 20
## mat:pat NA:Lineage A NA:Lineage B NA:mat
## 11 1215 322 1541
## NA:NA NA:pat pat:Lineage A pat:Lineage B
## 6981 2639 36 5
## pat:mat pat:NA pat:pat
## 59 229 105Summary of POE overlaps (00 = neither peak nor gene biased, 01 = gene unbiased, peak biased, 10 = gene biased, peak unbiased, 11 = both gene and peak show allelic bias)
##
## 00 01 10 11
## 12234 25 259 184Fishers exact test of enrichment (do genes showing allelic bias overlap with peaks showing allelic bias)?
##
## Fisher's Exact Test for Count Data
##
## data: data.frame(x = c(tab[4], tab[3]), y = c(tab[2], tab[1]))
## p-value < 2.2e-16
## alternative hypothesis: true odds ratio is greater than 1
## 95 percent confidence interval:
## 236.9345 Inf
## sample estimates:
## odds ratio
## 347.5206ASI: “allelic similarlty index”. Here, the axis values represent allelic transcription (x) and HPTM enrichment (y), with positive values being biased toward the paternal allele, and negative values being biased toward the maternal allele. ASI values are “=” for transcript and peak being baised toward the same allele, “NS” for either transcript or peak not showing allelic bias, and “≠” for biased toward opposite alleles.
QL
Gene:Peak (categories of allelic bias)
##
## Lineage A:Lineage A Lineage A:NA Lineage A:pat Lineage B:Lineage A
## 2 2 2 1
## Lineage B:mat Lineage B:NA Lineage B:pat mat:Lineage A
## 1 12 17 3
## mat:Lineage B mat:mat mat:NA mat:pat
## 1 2 50 31
## NA:Lineage A NA:Lineage B NA:mat NA:NA
## 759 338 1534 9161
## NA:pat pat:Lineage A pat:Lineage B pat:mat
## 5776 33 16 49
## pat:NA pat:pat
## 324 231Summary of overlaps (00 = neither peak nor gene biased, 01 = gene unbiased, peak biased, 10 = gene biased, peak unbiased, 11 = both gene and peak show allelic bias)
##
## 00 01 10 11
## 16843 55 366 313Fishers exact test of enrichment (do genes showing allelic bias overlap with peaks showing allelic bias)?
##
## Fisher's Exact Test for Count Data
##
## data: data.frame(x = c(tab[4], tab[3]), y = c(tab[2], tab[1]))
## p-value < 2.2e-16
## alternative hypothesis: true odds ratio is greater than 1
## 95 percent confidence interval:
## 199.8675 Inf
## sample estimates:
## odds ratio
## 260.0518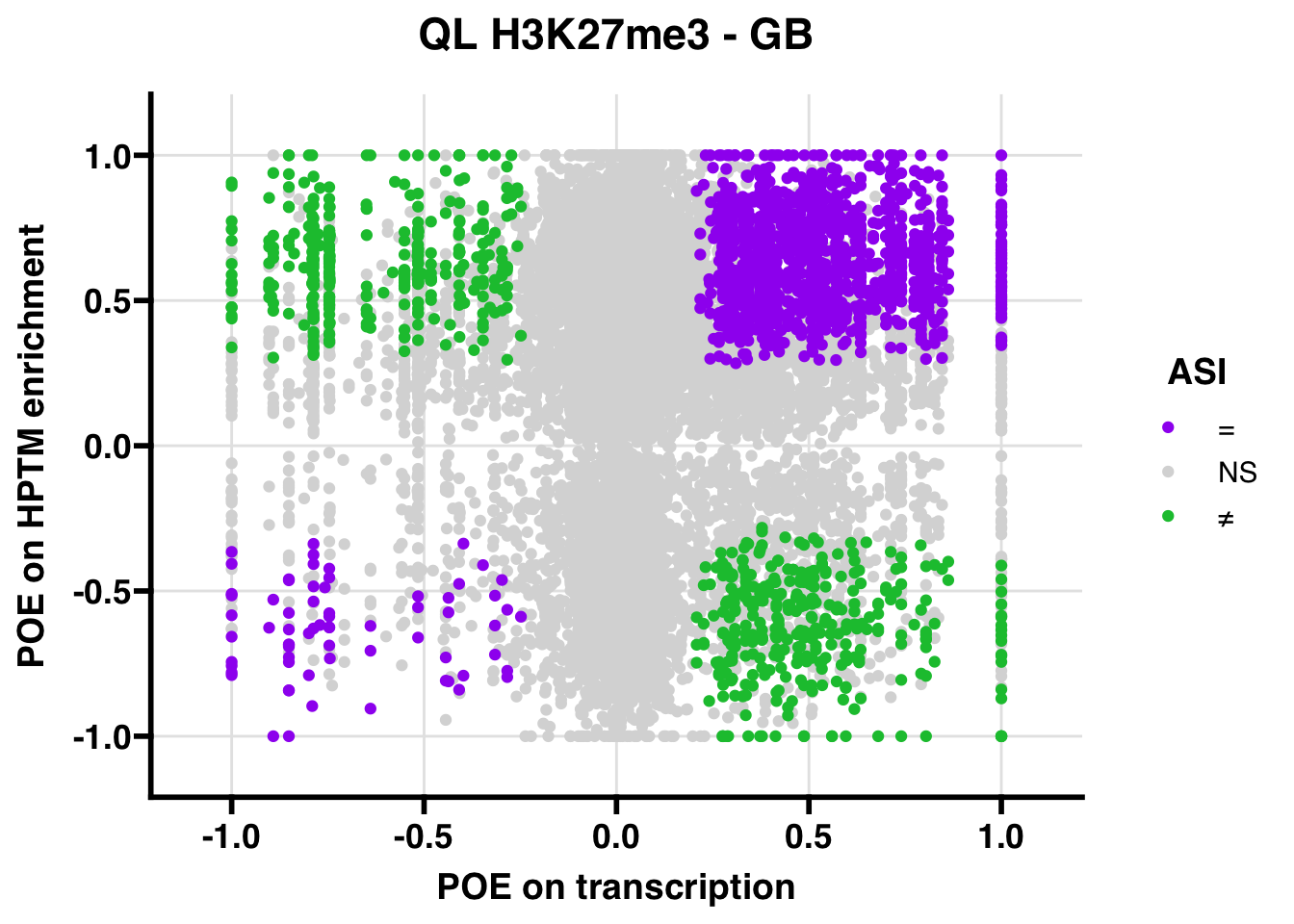
Promoter
WL
Gene:Peak (categories of allelic bias)
##
## Lineage A:Lineage A Lineage A:NA Lineage B:NA mat:mat
## 1 1 3 2
## mat:NA mat:pat NA:Lineage A NA:Lineage B
## 3 2 178 39
## NA:mat NA:NA NA:pat pat:Lineage A
## 217 1008 371 5
## pat:mat pat:NA pat:pat
## 8 31 18Summary of overlaps (00 = neither peak nor gene biased, 01 = gene unbiased, peak biased, 10 = gene biased, peak unbiased, 11 = both gene and peak show allelic bias)
##
## 00 01 10 11
## 1775 3 5 30Fishers exact test of enrichment (do genes showing allelic bias overlap with peaks showing allelic bias)?
##
## Fisher's Exact Test for Count Data
##
## data: data.frame(x = c(tab[4], tab[3]), y = c(tab[2], tab[1]))
## p-value < 2.2e-16
## alternative hypothesis: true odds ratio is greater than 1
## 95 percent confidence interval:
## 837.0586 Inf
## sample estimates:
## odds ratio
## 3190.375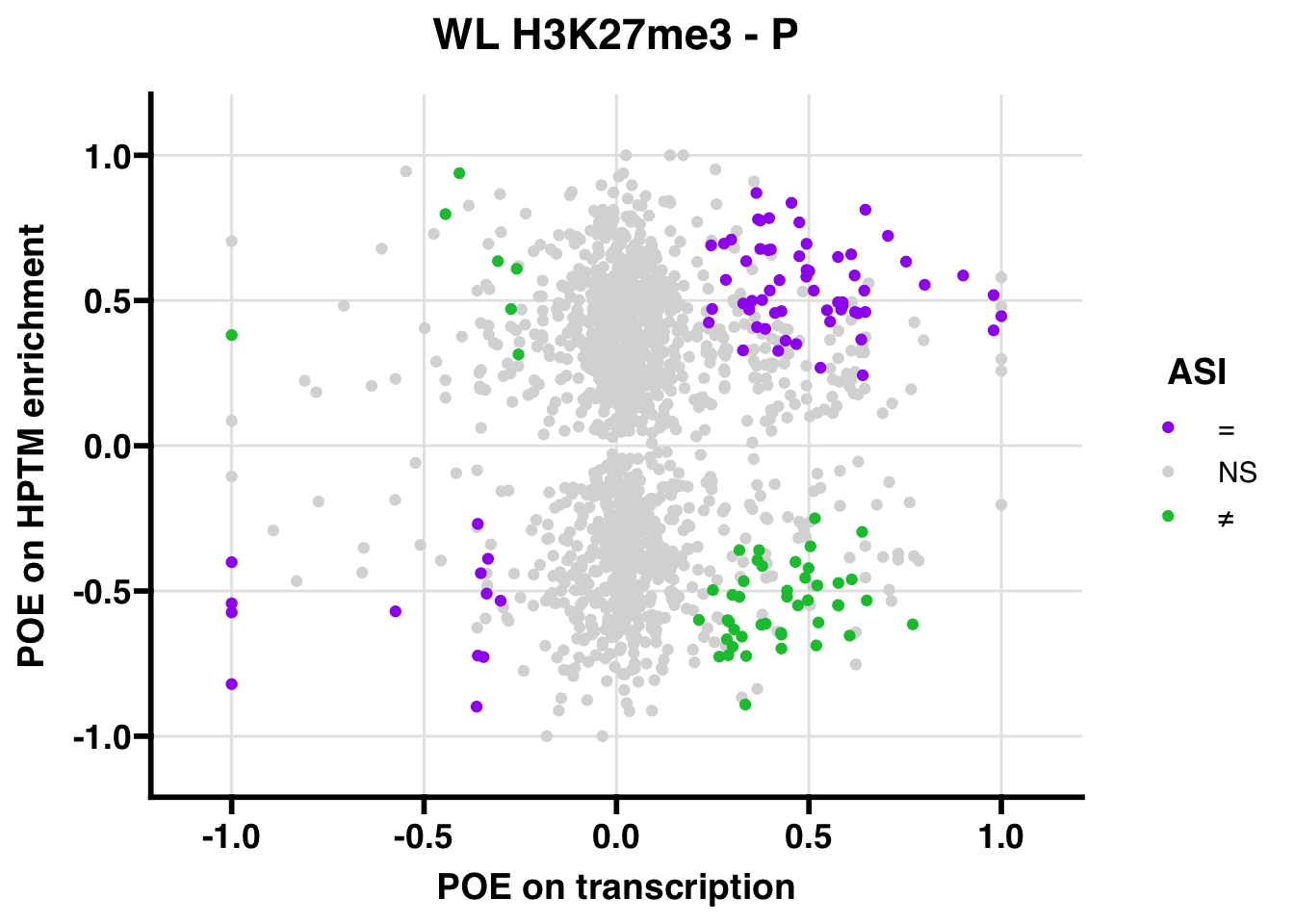
QL
Gene:Peak (categories of allelic bias)
##
## Lineage A:NA Lineage A:pat Lineage B:NA mat:Lineage B mat:NA
## 2 2 2 1 2
## mat:pat NA:Lineage A NA:Lineage B NA:mat NA:NA
## 2 118 37 187 1164
## NA:pat pat:Lineage A pat:mat pat:NA pat:pat
## 899 8 10 38 34Summary of overlaps (00 = neither peak nor gene biased, 01 = gene unbiased, peak biased, 10 = gene biased, peak unbiased, 11 = both gene and peak show allelic bias)
##
## 00 01 10 11
## 2341 11 12 46Fishers exact test of enrichment (do genes showing allelic bias overlap with peaks showing allelic bias)?
##
## Fisher's Exact Test for Count Data
##
## data: data.frame(x = c(tab[4], tab[3]), y = c(tab[2], tab[1]))
## p-value < 2.2e-16
## alternative hypothesis: true odds ratio is greater than 1
## 95 percent confidence interval:
## 358.0999 Inf
## sample estimates:
## odds ratio
## 798.7605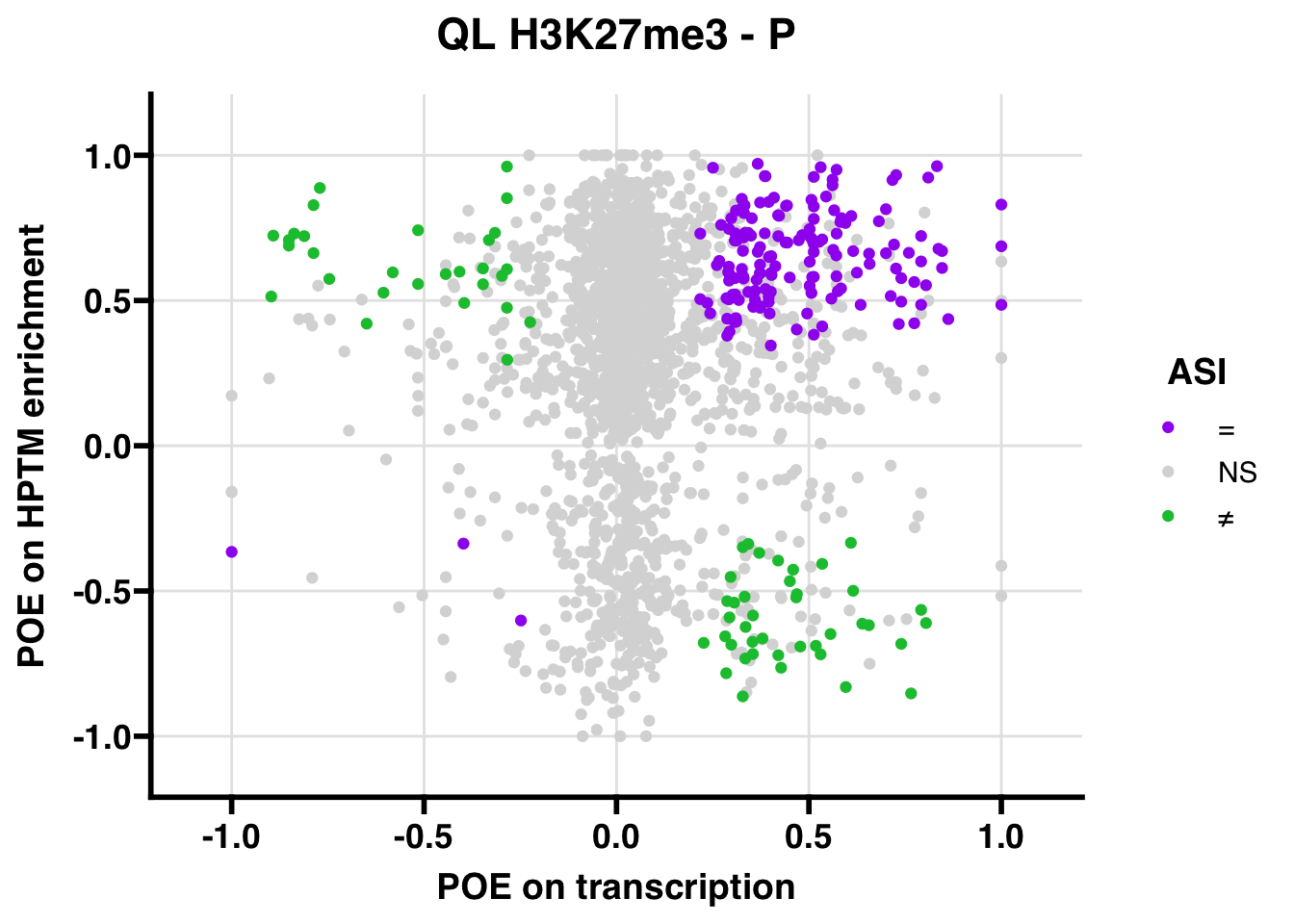
H3K27ac
Gene body
WL
Gene:Peak (categories of allelic bias)
##
## Lineage A:Lineage A Lineage A:NA Lineage A:pat Lineage B:Lineage A
## 2 7 1 7
## Lineage B:Lineage B Lineage B:mat Lineage B:NA Lineage B:pat
## 1 16 41 11
## mat:Lineage A mat:Lineage B mat:mat mat:NA
## 3 1 2 27
## mat:pat NA:Lineage A NA:Lineage B NA:mat
## 16 1086 247 1864
## NA:NA NA:pat pat:Lineage A pat:Lineage B
## 7627 2986 32 10
## pat:mat pat:NA pat:pat
## 63 233 120Summary of overlaps (00 = neither peak nor gene biased, 01 = gene unbiased, peak biased, 10 = gene biased, peak unbiased, 11 = both gene and peak show allelic bias)
##
## 00 01 10 11
## 13326 32 253 201Fishers exact test of enrichment (do genes showing allelic bias overlap with peaks showing allelic bias)?
##
## Fisher's Exact Test for Count Data
##
## data: data.frame(x = c(tab[4], tab[3]), y = c(tab[2], tab[1]))
## p-value < 2.2e-16
## alternative hypothesis: true odds ratio is greater than 1
## 95 percent confidence interval:
## 233.7337 Inf
## sample estimates:
## odds ratio
## 332.4454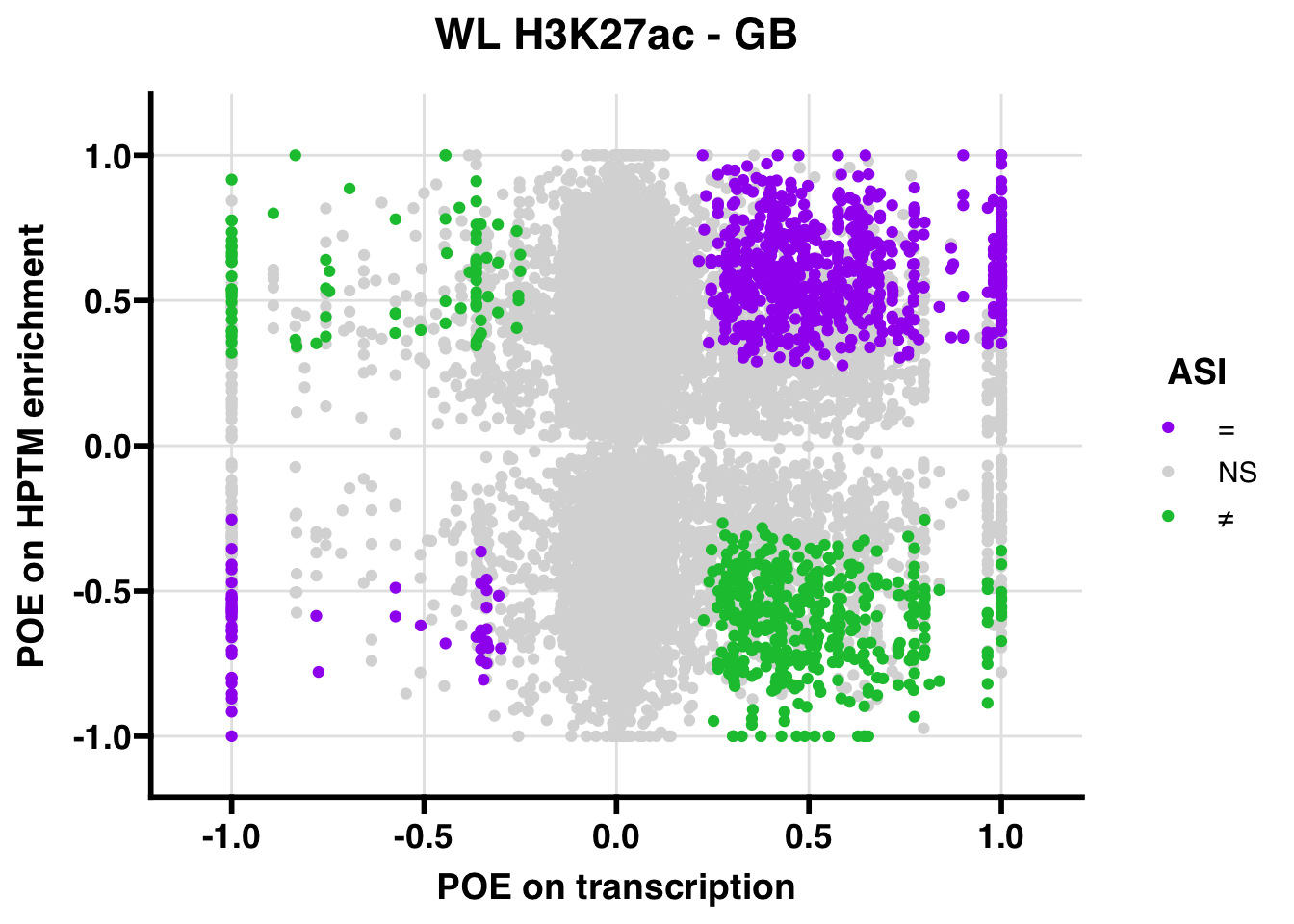
QL
Gene:Peak (categories of allelic bias)
##
## Lineage A:NA Lineage A:pat Lineage B:Lineage A Lineage B:Lineage B
## 2 5 1 1
## Lineage B:mat Lineage B:NA Lineage B:pat mat:Lineage A
## 1 18 16 5
## mat:Lineage B mat:mat mat:NA mat:pat
## 1 1 39 19
## NA:Lineage A NA:Lineage B NA:mat NA:NA
## 919 430 1333 8827
## NA:pat pat:Lineage A pat:Lineage B pat:mat
## 5847 31 24 48
## pat:NA pat:pat
## 320 250Summary of overlaps (00 = neither peak nor gene biased, 01 = gene unbiased, peak biased, 10 = gene biased, peak unbiased, 11 = both gene and peak show allelic bias)
##
## 00 01 10 11
## 16609 69 366 318Fishers exact test of enrichment (do genes showing allelic bias overlap with peaks showing allelic bias)?
##
## Fisher's Exact Test for Count Data
##
## data: data.frame(x = c(tab[4], tab[3]), y = c(tab[2], tab[1]))
## p-value < 2.2e-16
## alternative hypothesis: true odds ratio is greater than 1
## 95 percent confidence interval:
## 164.6826 Inf
## sample estimates:
## odds ratio
## 208.1453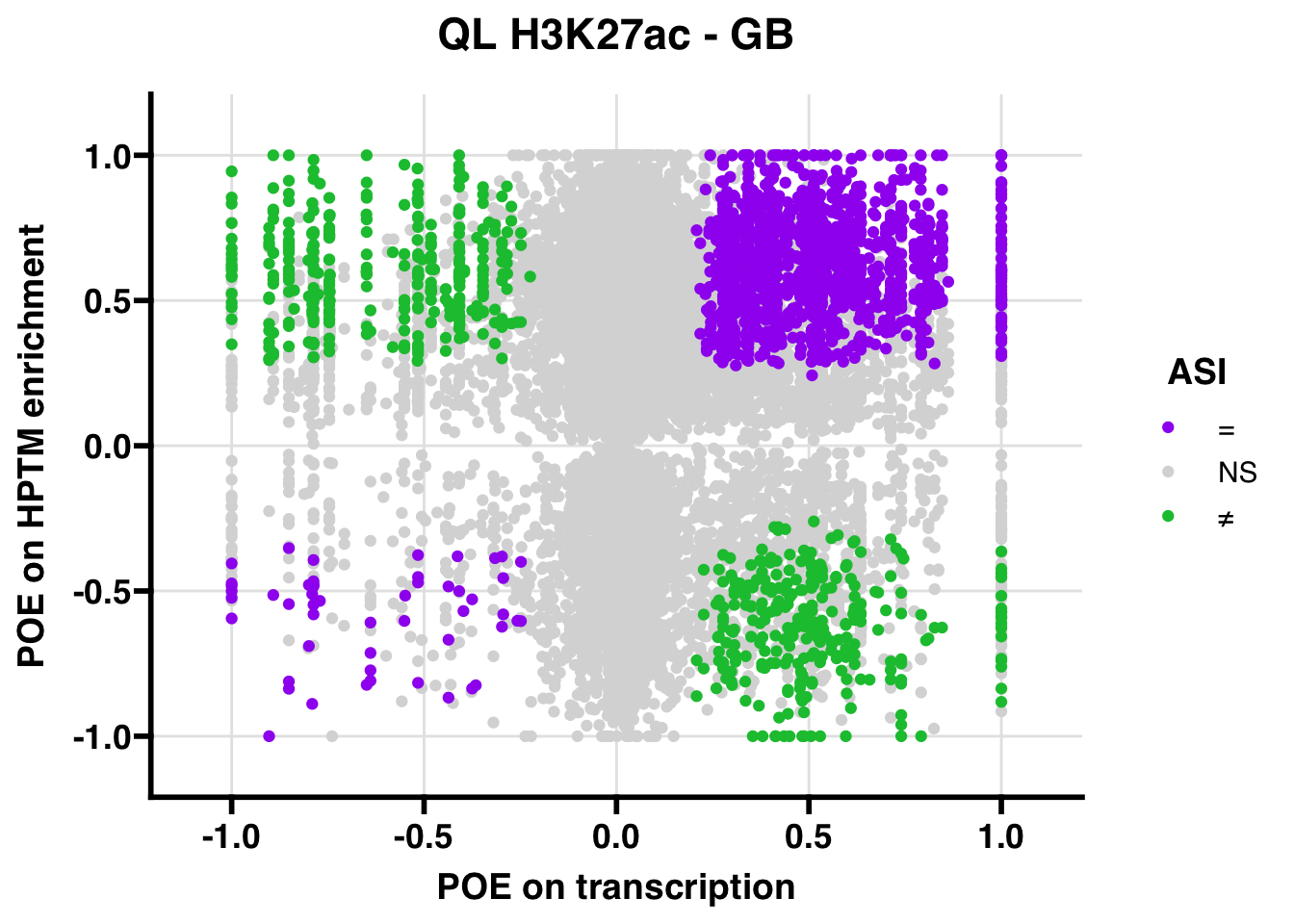
Promoter
WL
Gene:Peak (categories of allelic bias)
##
## Lineage A:Lineage A Lineage A:NA Lineage B:NA Lineage B:pat
## 1 1 3 2
## mat:mat mat:NA mat:pat NA:Lineage A
## 2 3 1 180
## NA:Lineage B NA:mat NA:NA NA:pat
## 38 265 1256 482
## pat:Lineage A pat:Lineage B pat:mat pat:NA
## 3 3 9 40
## pat:pat
## 20Summary of overlaps (00 = neither peak nor gene biased, 01 = gene unbiased, peak biased, 10 = gene biased, peak unbiased, 11 = both gene and peak show allelic bias)
##
## 00 01 10 11
## 2176 7 7 32Fishers exact test of enrichment (do genes showing allelic bias overlap with peaks showing allelic bias)?
##
## Fisher's Exact Test for Count Data
##
## data: data.frame(x = c(tab[4], tab[3]), y = c(tab[2], tab[1]))
## p-value < 2.2e-16
## alternative hypothesis: true odds ratio is greater than 1
## 95 percent confidence interval:
## 488.4372 Inf
## sample estimates:
## odds ratio
## 1342.753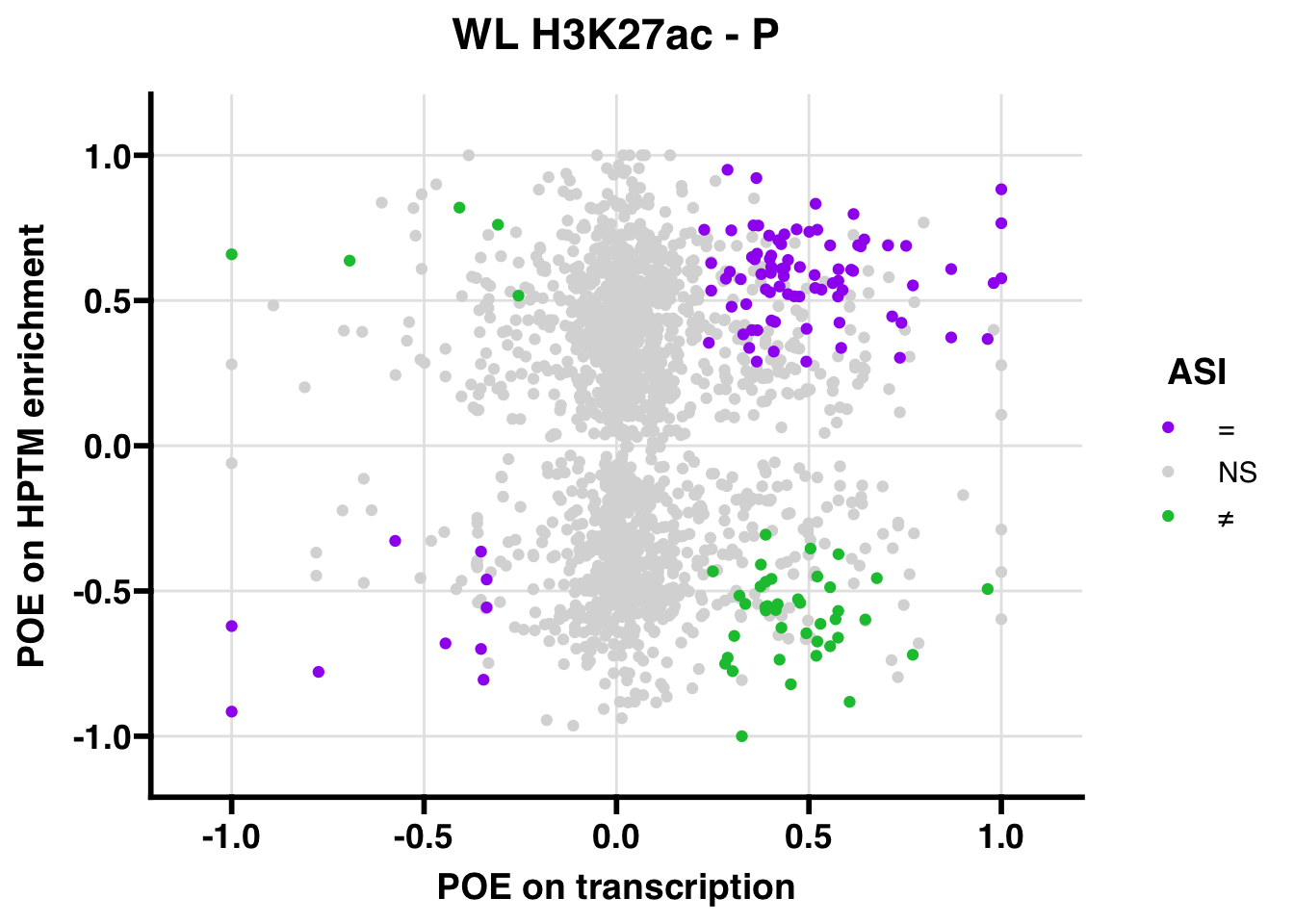
QL
Gene:Peak (categories of allelic bias)
##
## Lineage A:NA Lineage A:pat Lineage B:NA Lineage B:pat mat:Lineage B
## 1 3 1 1 1
## mat:NA mat:pat NA:Lineage A NA:Lineage B NA:mat
## 2 1 113 56 192
## NA:NA NA:pat pat:Lineage A pat:Lineage B pat:mat
## 1219 859 2 2 6
## pat:NA pat:pat
## 42 33Summary of overlaps (00 = neither peak nor gene biased, 01 = gene unbiased, peak biased, 10 = gene biased, peak unbiased, 11 = both gene and peak show allelic bias)
##
## 00 01 10 11
## 2380 11 12 40Fishers exact test of enrichment (do genes showing allelic bias overlap with peaks showing allelic bias)?
##
## Fisher's Exact Test for Count Data
##
## data: data.frame(x = c(tab[4], tab[3]), y = c(tab[2], tab[1]))
## p-value < 2.2e-16
## alternative hypothesis: true odds ratio is greater than 1
## 95 percent confidence interval:
## 313.8559 Inf
## sample estimates:
## odds ratio
## 700.1813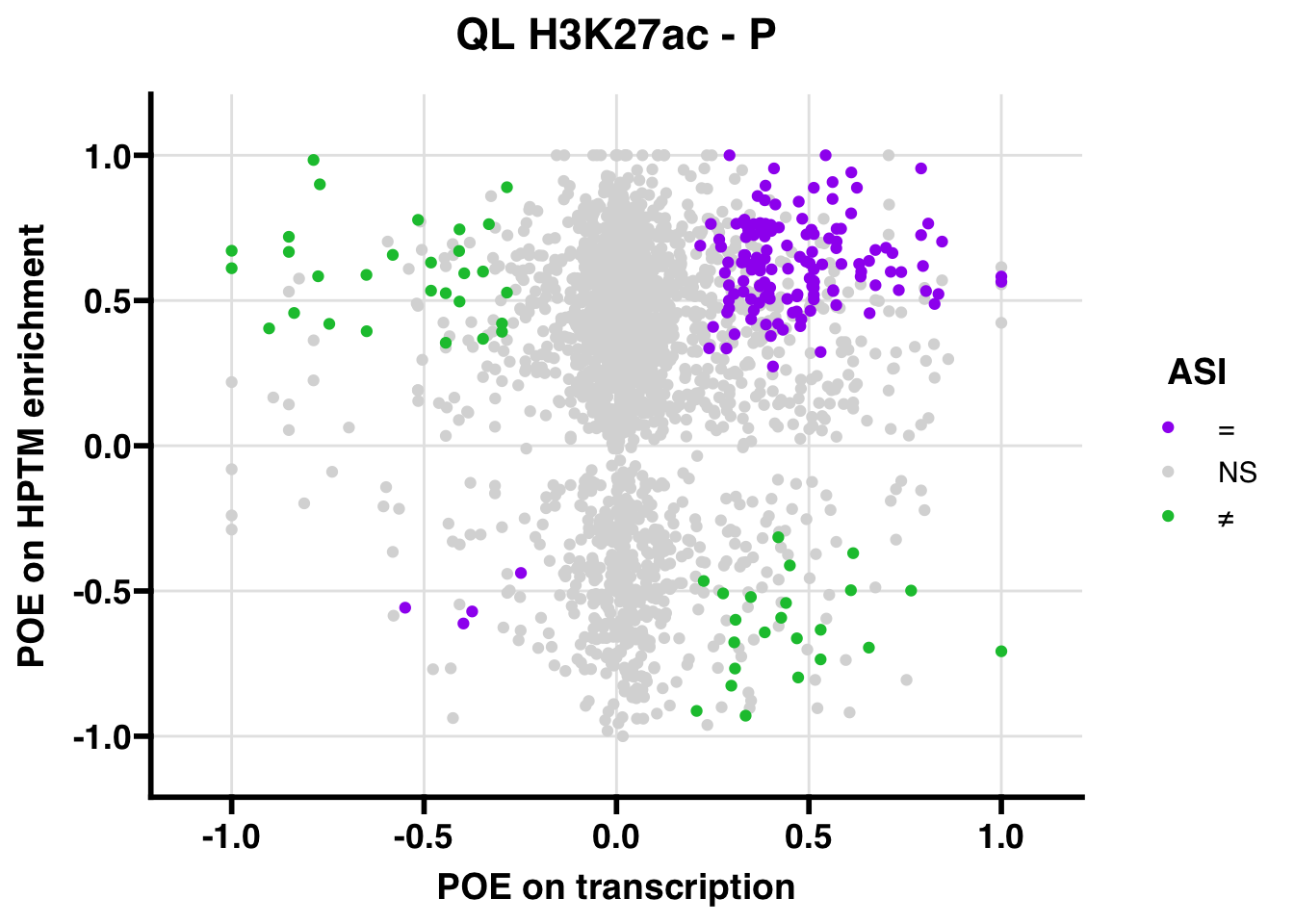
H3K4me3
Gene body
WL
Gene:Peak (categories of allelic bias)
##
## Lineage A:Lineage A Lineage A:NA Lineage A:pat Lineage B:Lineage A
## 2 5 4 3
## Lineage B:Lineage B Lineage B:mat Lineage B:NA Lineage B:pat
## 1 13 32 9
## mat:Lineage A mat:mat mat:NA mat:pat
## 2 6 21 12
## NA:Lineage A NA:Lineage B NA:mat NA:NA
## 539 128 1786 7420
## NA:pat pat:Lineage A pat:Lineage B pat:mat
## 2843 14 6 48
## pat:NA pat:pat
## 213 110Summary of overlaps (00 = neither peak nor gene biased, 01 = gene unbiased, peak biased, 10 = gene biased, peak unbiased, 11 = both gene and peak show allelic bias)
##
## 00 01 10 11
## 12322 14 208 176Fishers exact test of enrichment (do genes showing allelic bias overlap with peaks showing allelic bias)?
##
## Fisher's Exact Test for Count Data
##
## data: data.frame(x = c(tab[4], tab[3]), y = c(tab[2], tab[1]))
## p-value < 2.2e-16
## alternative hypothesis: true odds ratio is greater than 1
## 95 percent confidence interval:
## 459.8188 Inf
## sample estimates:
## odds ratio
## 741.8472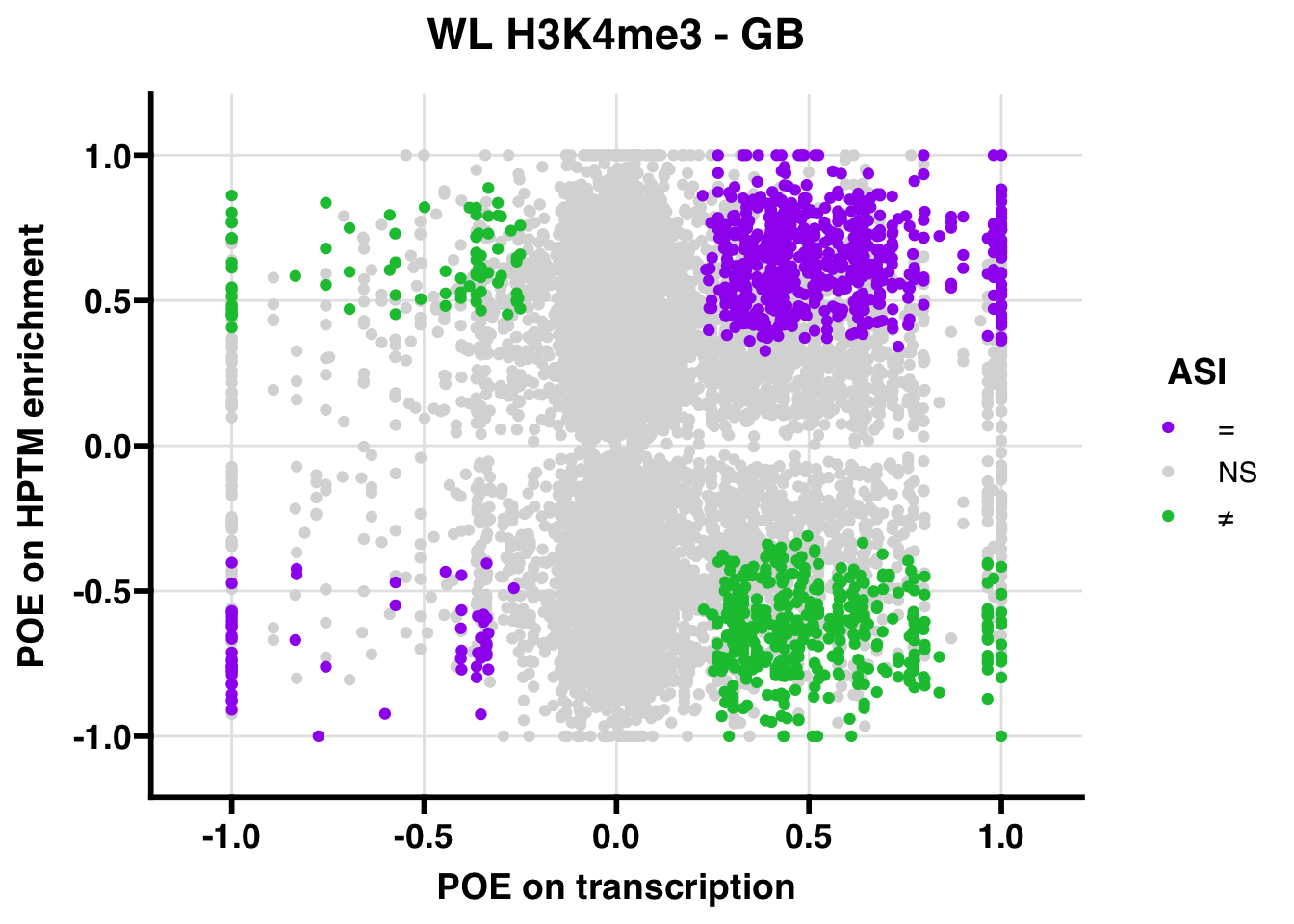
QL
Gene:Peak (categories of allelic bias)
##
## Lineage A:Lineage A Lineage A:NA Lineage A:pat Lineage B:Lineage B
## 1 3 4 1
## Lineage B:mat Lineage B:NA Lineage B:pat mat:Lineage A
## 2 15 9 5
## mat:Lineage B mat:mat mat:NA mat:pat
## 1 4 28 26
## NA:Lineage A NA:Lineage B NA:mat NA:NA
## 609 272 1237 8025
## NA:pat pat:Lineage A pat:Lineage B pat:mat
## 4844 25 22 43
## pat:NA pat:pat
## 297 224Summary of overlaps (00 = neither peak nor gene biased, 01 = gene unbiased, peak biased, 10 = gene biased, peak unbiased, 11 = both gene and peak show allelic bias)
##
## 00 01 10 11
## 14332 41 329 297Fishers exact test of enrichment (do genes showing allelic bias overlap with peaks showing allelic bias)?
##
## Fisher's Exact Test for Count Data
##
## data: data.frame(x = c(tab[4], tab[3]), y = c(tab[2], tab[1]))
## p-value < 2.2e-16
## alternative hypothesis: true odds ratio is greater than 1
## 95 percent confidence interval:
## 235.3106 Inf
## sample estimates:
## odds ratio
## 315.9013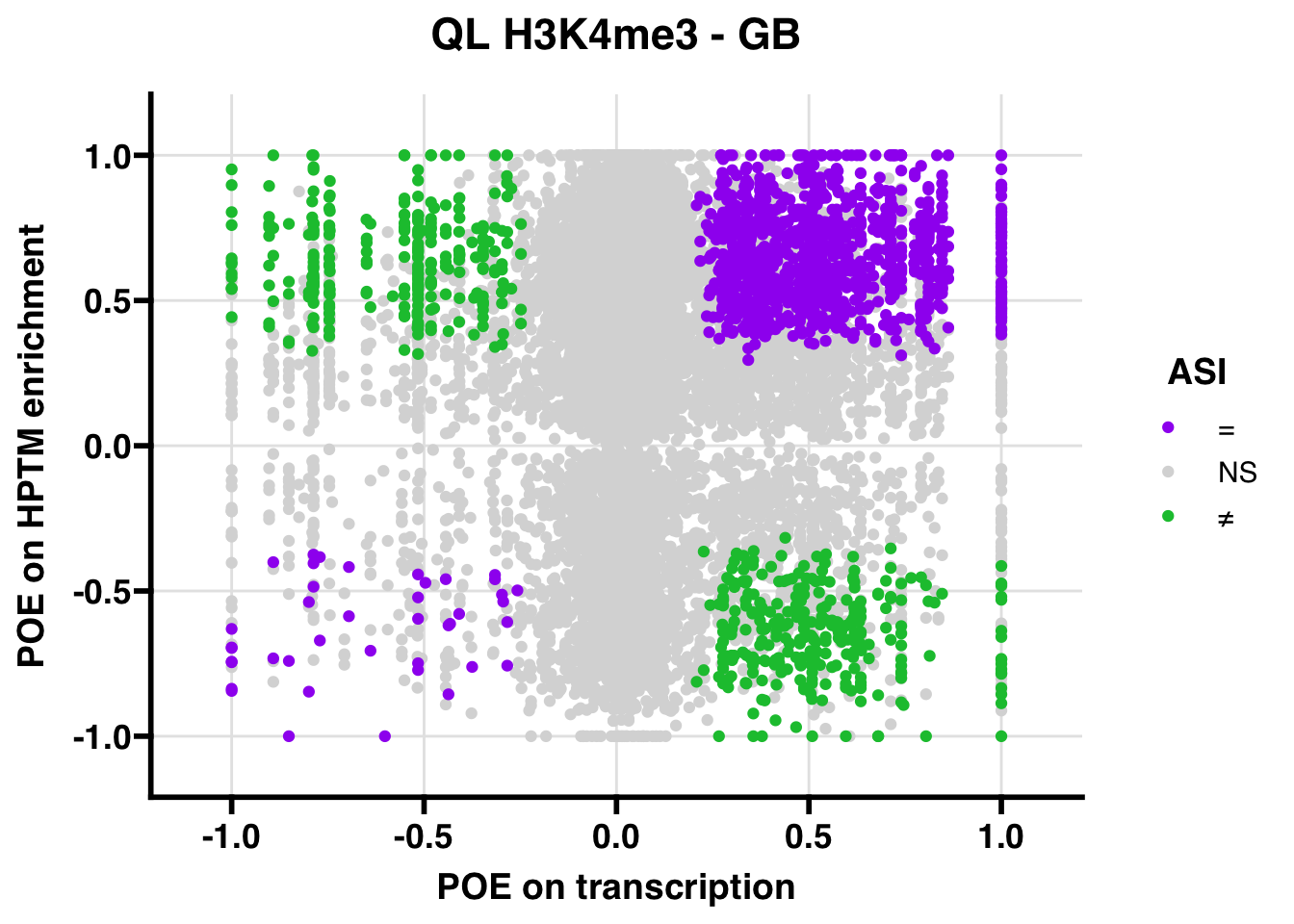
Promoter
WL
Gene:Peak (categories of allelic bias)
##
## Lineage A:Lineage A Lineage A:NA Lineage A:pat Lineage B:NA
## 1 1 1 1
## Lineage B:pat mat:mat mat:NA mat:pat
## 3 1 2 1
## NA:Lineage A NA:Lineage B NA:mat NA:NA
## 94 17 267 1104
## NA:pat pat:Lineage A pat:Lineage B pat:mat
## 465 3 1 8
## pat:NA pat:pat
## 33 26Summary of overlaps (00 = neither peak nor gene biased, 01 = gene unbiased, peak biased, 10 = gene biased, peak unbiased, 11 = both gene and peak show allelic bias)
##
## 00 01 10 11
## 1907 3 3 36Fishers exact test of enrichment (do genes showing allelic bias overlap with peaks showing allelic bias)?
##
## Fisher's Exact Test for Count Data
##
## data: data.frame(x = c(tab[4], tab[3]), y = c(tab[2], tab[1]))
## p-value < 2.2e-16
## alternative hypothesis: true odds ratio is greater than 1
## 95 percent confidence interval:
## 1602.424 Inf
## sample estimates:
## odds ratio
## 7569.223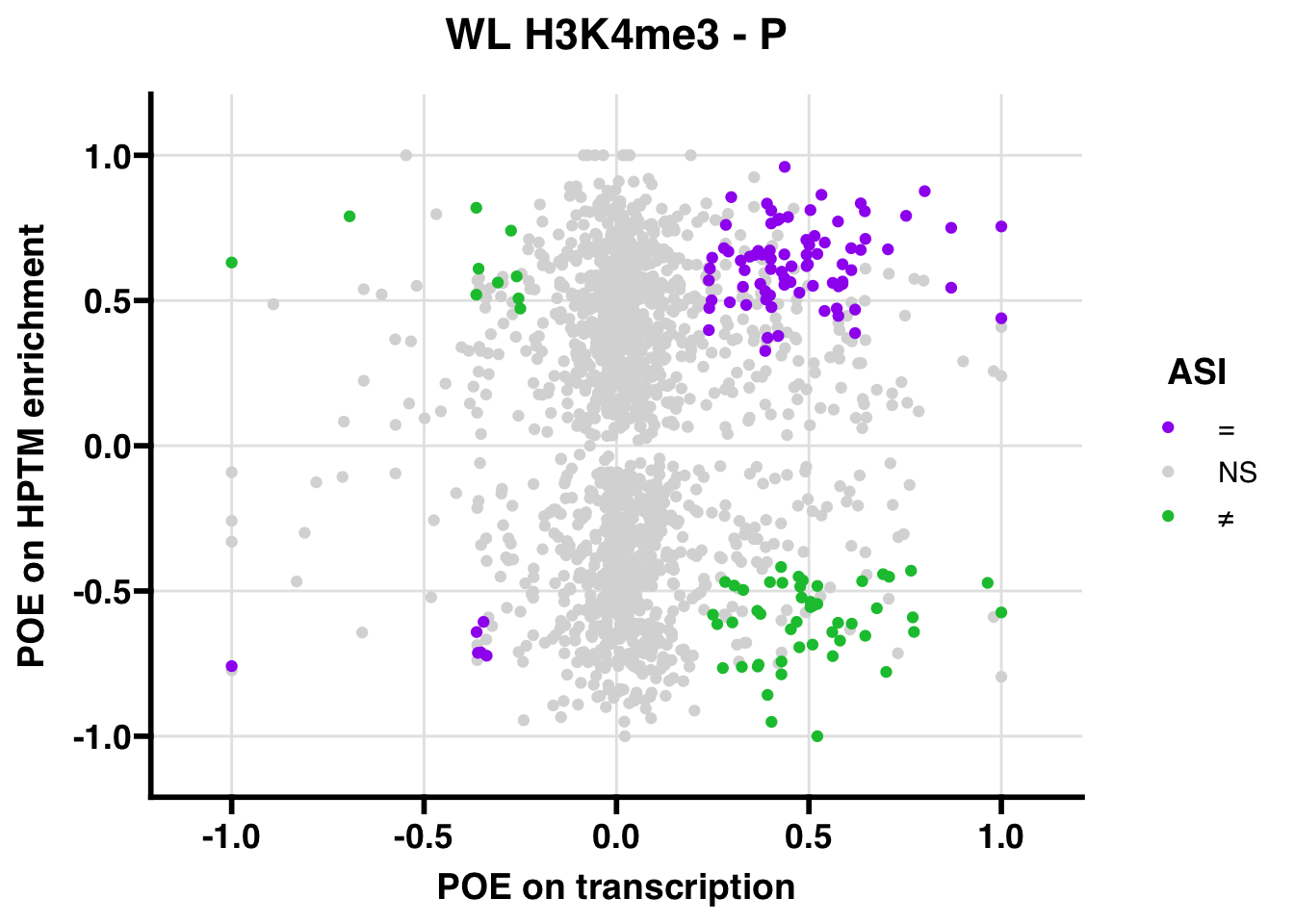
QL
Gene:Peak (categories of allelic bias)
##
## Lineage A:NA Lineage A:pat Lineage B:pat mat:Lineage B mat:mat
## 3 3 2 1 2
## mat:NA NA:Lineage A NA:Lineage B NA:mat NA:NA
## 2 74 36 167 1075
## NA:pat pat:Lineage A pat:Lineage B pat:mat pat:NA
## 697 1 3 3 44
## pat:pat
## 35Summary of overlaps (00 = neither peak nor gene biased, 01 = gene unbiased, peak biased, 10 = gene biased, peak unbiased, 11 = both gene and peak show allelic bias)
##
## 00 01 10 11
## 1998 6 9 40Fishers exact test of enrichment (do genes showing allelic bias overlap with peaks showing allelic bias)?
##
## Fisher's Exact Test for Count Data
##
## data: data.frame(x = c(tab[4], tab[3]), y = c(tab[2], tab[1]))
## p-value < 2.2e-16
## alternative hypothesis: true odds ratio is greater than 1
## 95 percent confidence interval:
## 521.7834 Inf
## sample estimates:
## odds ratio
## 1346.678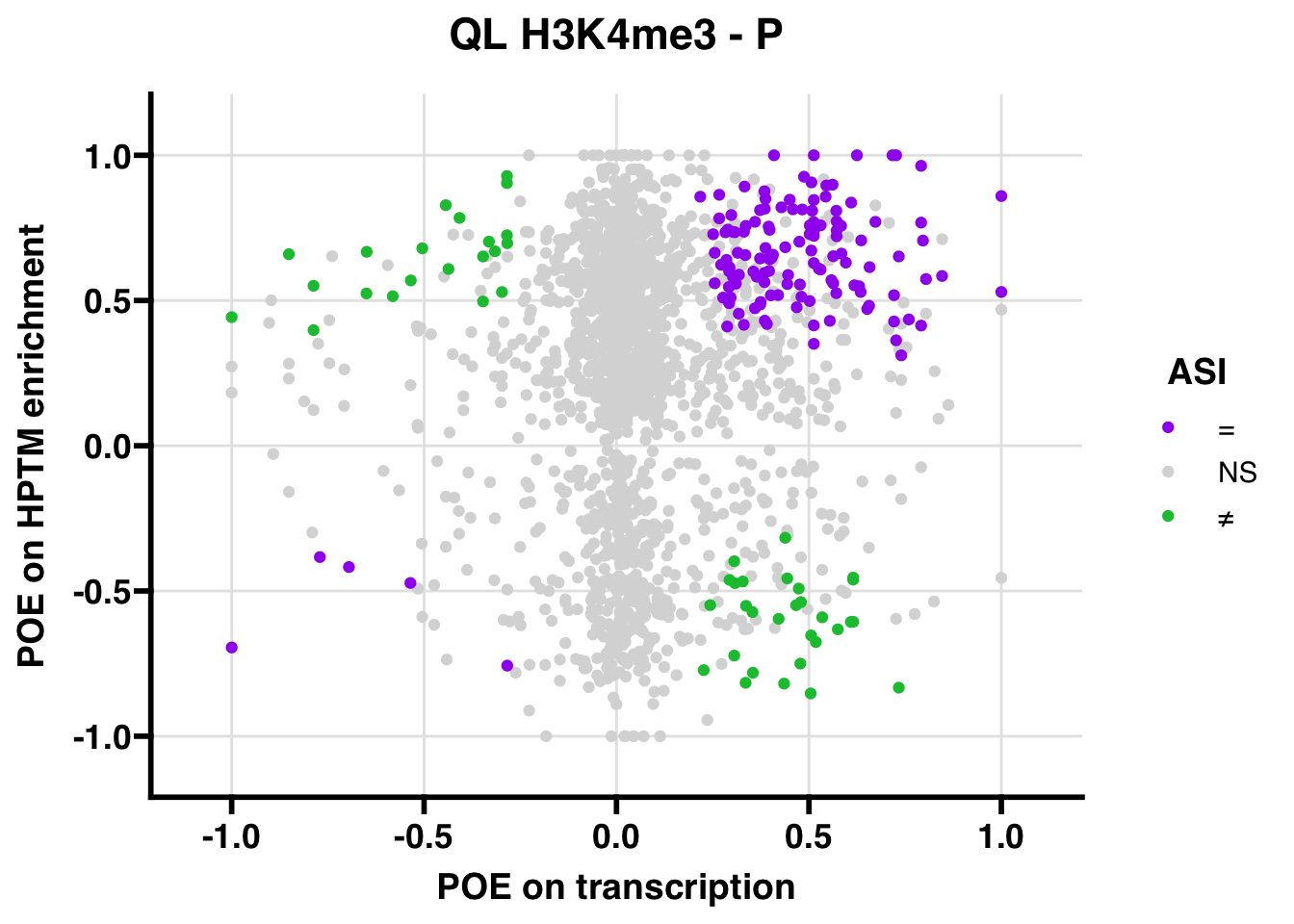
7.2 Block 2
H3K27me3
Gene body
WL
Gene:Peak (categories of allelic bias)
##
## Lineage B:Lineage A Lineage B:mat Lineage B:NA Lineage B:pat
## 1 1 11 3
## mat:Lineage A mat:Lineage B mat:mat mat:NA
## 4 3 19 54
## mat:pat NA:Lineage A NA:Lineage B NA:mat
## 10 569 727 1436
## NA:NA NA:pat pat:Lineage A pat:Lineage B
## 5053 1949 6 17
## pat:mat pat:NA pat:pat
## 37 83 29Summary of overlaps (00 = neither peak nor gene biased, 01 = gene unbiased, peak biased, 10 = gene biased, peak unbiased, 11 = both gene and peak show allelic bias)
##
## 00 01 10 11
## 9474 23 145 95Fishers exact test of enrichment (do genes showing allelic bias overlap with peaks showing allelic bias)?
##
## Fisher's Exact Test for Count Data
##
## data: data.frame(x = c(tab[4], tab[3]), y = c(tab[2], tab[1]))
## p-value < 2.2e-16
## alternative hypothesis: true odds ratio is greater than 1
## 95 percent confidence interval:
## 175.8221 Inf
## sample estimates:
## odds ratio
## 269.862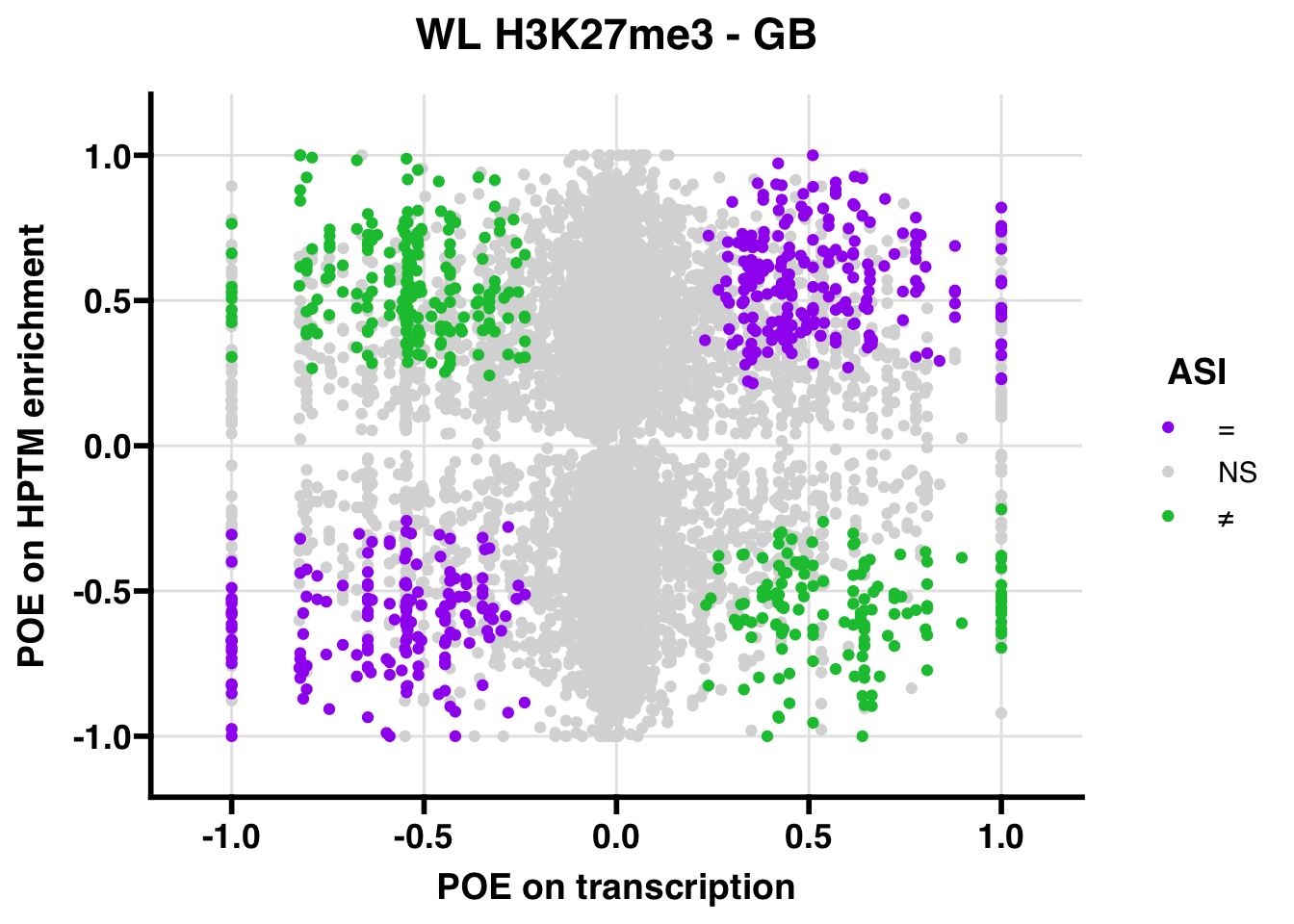
QL
Gene:Peak (categories of allelic bias)
##
## Lineage B:Lineage A Lineage B:Lineage B Lineage B:mat Lineage B:NA
## 1 2 3 7
## Lineage B:pat mat:Lineage A mat:Lineage B mat:mat
## 6 1 1 1
## mat:NA mat:pat NA:Lineage A NA:Lineage B
## 15 6 563 617
## NA:mat NA:NA NA:pat pat:Lineage A
## 941 5414 3362 3
## pat:Lineage B pat:mat pat:NA pat:pat
## 8 10 84 73Summary of overlaps (00 = neither peak nor gene biased, 01 = gene unbiased, peak biased, 10 = gene biased, peak unbiased, 11 = both gene and peak show allelic bias)
##
## 00 01 10 11
## 10691 26 98 90Fishers exact test of enrichment (do genes showing allelic bias overlap with peaks showing allelic bias)?
##
## Fisher's Exact Test for Count Data
##
## data: data.frame(x = c(tab[4], tab[3]), y = c(tab[2], tab[1]))
## p-value < 2.2e-16
## alternative hypothesis: true odds ratio is greater than 1
## 95 percent confidence interval:
## 245.3571 Inf
## sample estimates:
## odds ratio
## 374.0261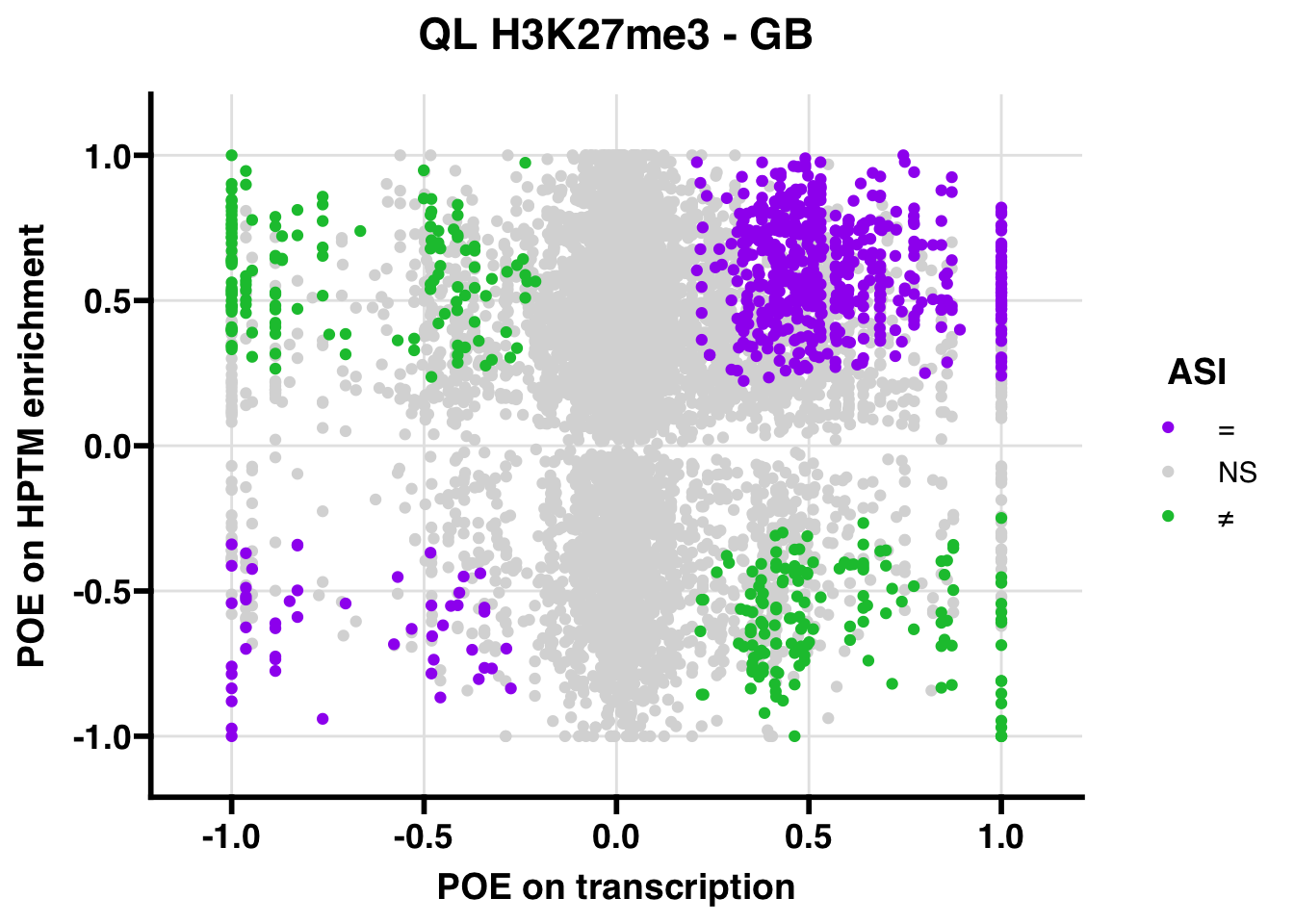
Promoter
WL
Gene:Peak (categories of allelic bias)
##
## mat:mat mat:NA mat:pat NA:Lineage A NA:Lineage B
## 1 9 1 72 79
## NA:mat NA:NA NA:pat pat:Lineage A pat:Lineage B
## 220 763 249 1 1
## pat:mat pat:NA pat:pat
## 2 11 3Summary of overlaps (00 = neither peak nor gene biased, 01 = gene unbiased, peak biased, 10 = gene biased, peak unbiased, 11 = both gene and peak show allelic bias)
##
## 00 01 11
## 1376 3 7Fishers exact test of enrichment (do genes showing allelic bias overlap with peaks showing allelic bias)?
## [1] "NA"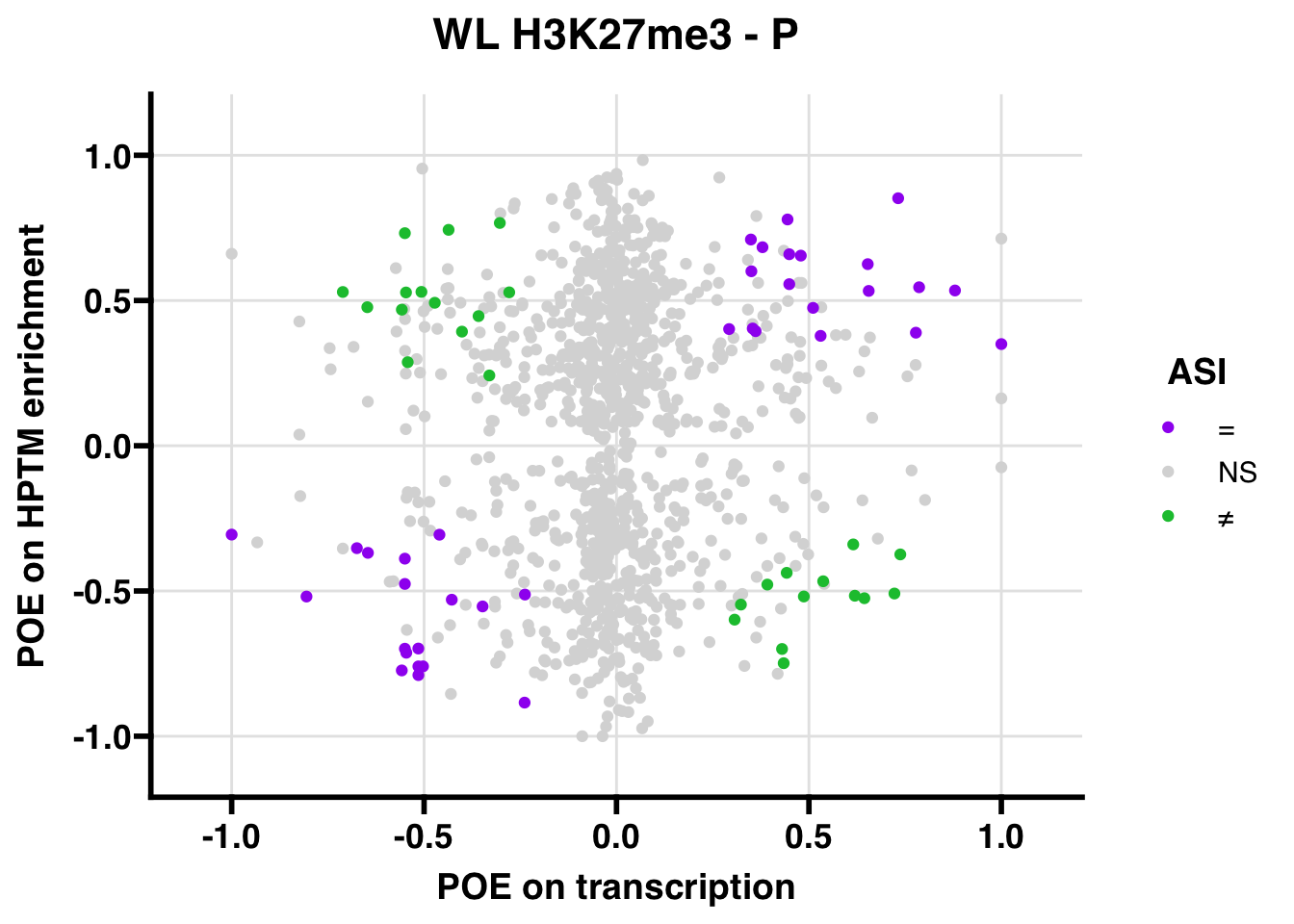
QL
Gene:Peak (categories of allelic bias)
##
## Lineage B:NA Lineage B:pat mat:NA mat:pat NA:Lineage A
## 1 1 3 3 86
## NA:Lineage B NA:mat NA:NA NA:pat pat:Lineage A
## 99 129 865 561 1
## pat:Lineage B pat:NA pat:pat
## 3 20 16Summary of overlaps (00 = neither peak nor gene biased, 01 = gene unbiased, peak biased, 10 = gene biased, peak unbiased, 11 = both gene and peak show allelic bias)
##
## 00 01 10 11
## 1720 2 4 19Fishers exact test of enrichment (do genes showing allelic bias overlap with peaks showing allelic bias)?
##
## Fisher's Exact Test for Count Data
##
## data: data.frame(x = c(tab[4], tab[3]), y = c(tab[2], tab[1]))
## p-value < 2.2e-16
## alternative hypothesis: true odds ratio is greater than 1
## 95 percent confidence interval:
## 721.7098 Inf
## sample estimates:
## odds ratio
## 3533.803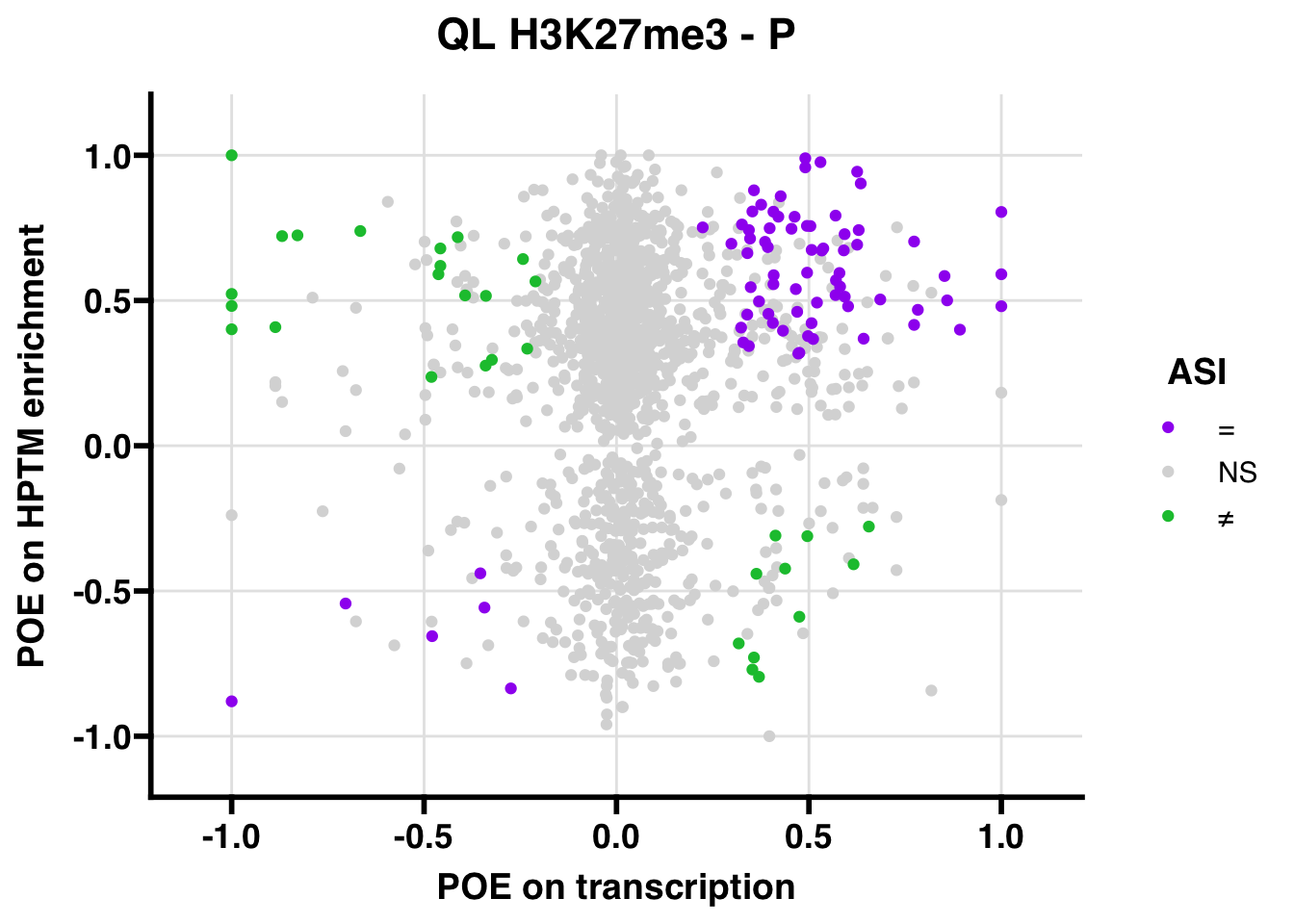
H3K27ac
Gene body
WL
Gene:Peak (categories of allelic bias)
##
## Lineage B:Lineage A Lineage B:mat Lineage B:NA Lineage B:pat
## 1 2 6 3
## mat:Lineage A mat:Lineage B mat:mat mat:NA
## 3 6 10 41
## mat:pat NA:Lineage A NA:Lineage B NA:mat
## 9 282 336 843
## NA:NA NA:pat pat:Lineage A pat:Lineage B
## 2933 1191 2 9
## pat:mat pat:NA pat:pat
## 24 40 28Summary of overlaps (00 = neither peak nor gene biased, 01 = gene unbiased, peak biased, 10 = gene biased, peak unbiased, 11 = both gene and peak show allelic bias)
##
## 00 01 10 11
## 5415 16 85 71Fishers exact test of enrichment (do genes showing allelic bias overlap with peaks showing allelic bias)?
##
## Fisher's Exact Test for Count Data
##
## data: data.frame(x = c(tab[4], tab[3]), y = c(tab[2], tab[1]))
## p-value < 2.2e-16
## alternative hypothesis: true odds ratio is greater than 1
## 95 percent confidence interval:
## 167.0016 Inf
## sample estimates:
## odds ratio
## 281.5192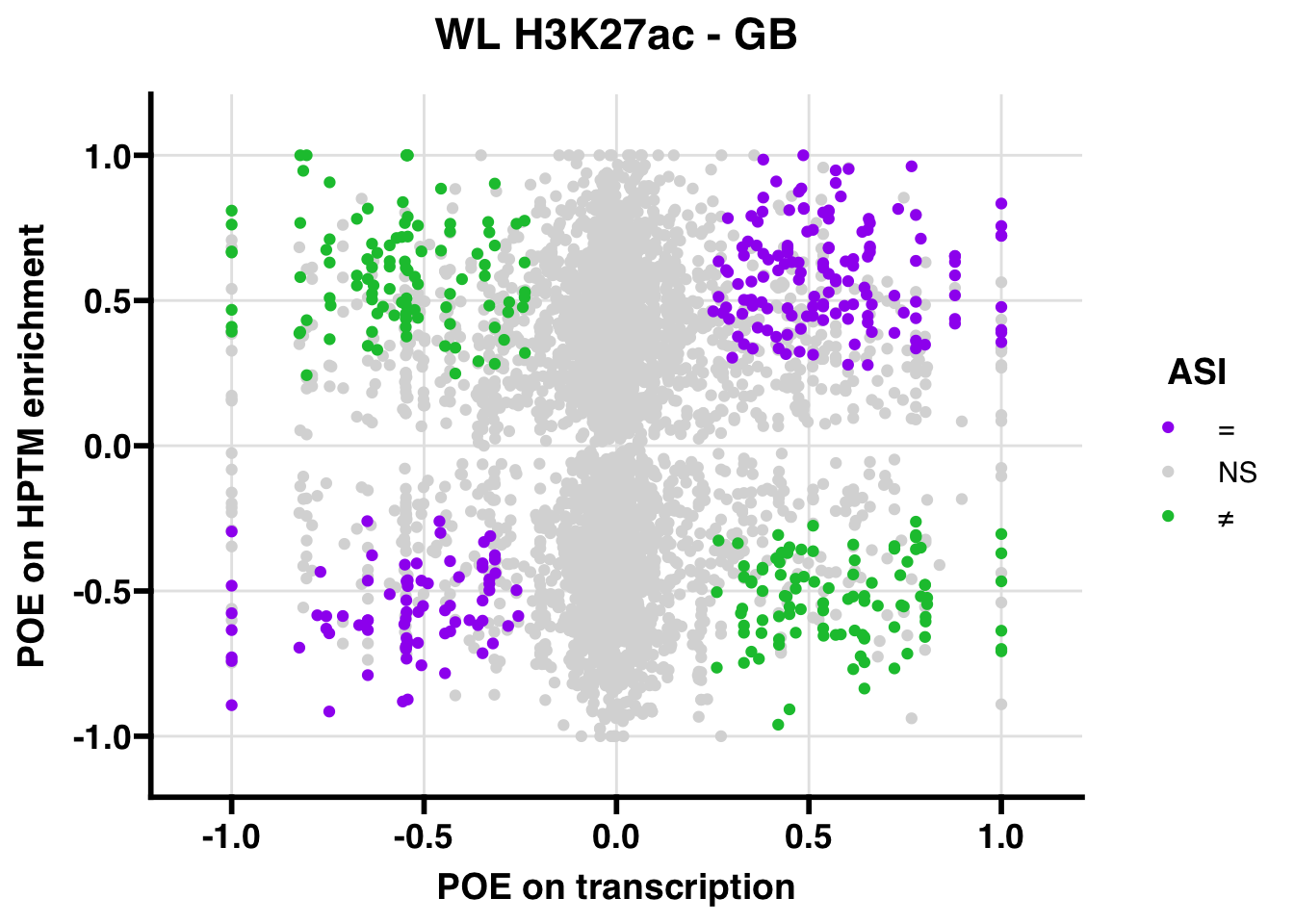
QL
Gene:Peak (categories of allelic bias)
##
## Lineage B:Lineage A Lineage B:mat Lineage B:NA Lineage B:pat
## 6 2 17 5
## mat:Lineage A mat:Lineage B mat:mat mat:NA
## 3 2 2 25
## mat:pat NA:Lineage A NA:Lineage B NA:mat
## 17 605 613 950
## NA:NA NA:pat pat:Lineage A pat:Lineage B
## 5427 3388 8 5
## pat:mat pat:NA pat:pat
## 8 64 84Summary of overlaps (00 = neither peak nor gene biased, 01 = gene unbiased, peak biased, 10 = gene biased, peak unbiased, 11 = both gene and peak show allelic bias)
##
## 00 01 10 11
## 10757 32 93 111Fishers exact test of enrichment (do genes showing allelic bias overlap with peaks showing allelic bias)?
##
## Fisher's Exact Test for Count Data
##
## data: data.frame(x = c(tab[4], tab[3]), y = c(tab[2], tab[1]))
## p-value < 2.2e-16
## alternative hypothesis: true odds ratio is greater than 1
## 95 percent confidence interval:
## 269.4737 Inf
## sample estimates:
## odds ratio
## 397.5212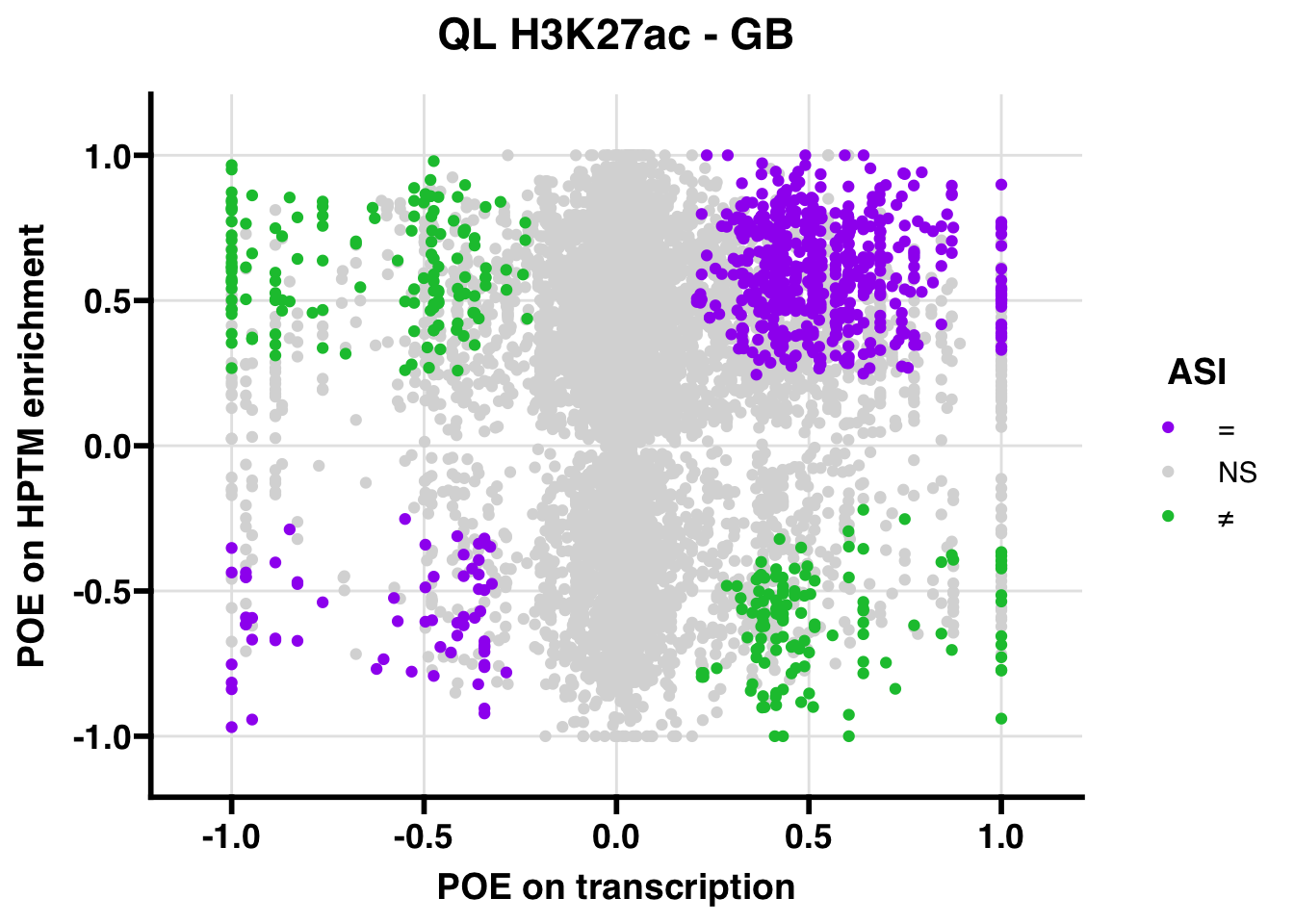
Promoter
WL
Gene:Peak (categories of allelic bias)
##
## mat:mat mat:NA mat:pat NA:Lineage A NA:Lineage B NA:mat
## 2 4 3 59 63 156
## NA:NA NA:pat pat:mat pat:NA pat:pat
## 569 192 3 9 3Summary of overlaps (00 = neither peak nor gene biased, 01 = gene unbiased, peak biased, 10 = gene biased, peak unbiased, 11 = both gene and peak show allelic bias)
##
## 00 01 10 11
## 1022 4 3 11Fishers exact test of enrichment (do genes showing allelic bias overlap with peaks showing allelic bias)?
##
## Fisher's Exact Test for Count Data
##
## data: data.frame(x = c(tab[4], tab[3]), y = c(tab[2], tab[1]))
## p-value < 2.2e-16
## alternative hypothesis: true odds ratio is greater than 1
## 95 percent confidence interval:
## 189.0323 Inf
## sample estimates:
## odds ratio
## 830.8785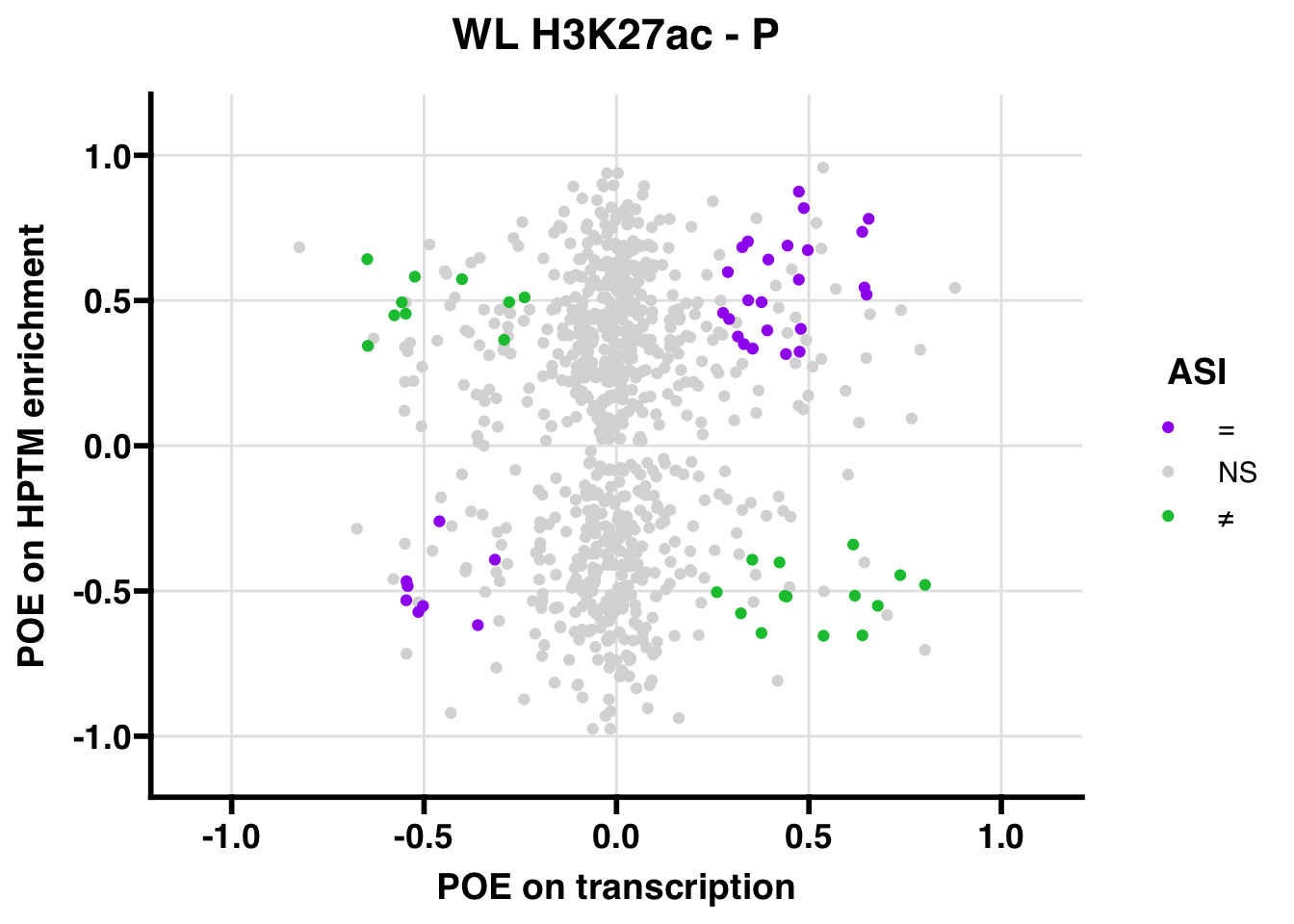
QL
Gene:Peak (categories of allelic bias)
##
## Lineage B:pat mat:NA mat:pat NA:Lineage A NA:Lineage B
## 1 1 5 99 92
## NA:mat NA:NA NA:pat pat:Lineage A pat:Lineage B
## 157 875 568 2 1
## pat:NA pat:pat
## 16 22Summary of overlaps (00 = neither peak nor gene biased, 01 = gene unbiased, peak biased, 10 = gene biased, peak unbiased, 11 = both gene and peak show allelic bias)
##
## 00 01 10 11
## 1762 4 3 27Fishers exact test of enrichment (do genes showing allelic bias overlap with peaks showing allelic bias)?
##
## Fisher's Exact Test for Count Data
##
## data: data.frame(x = c(tab[4], tab[3]), y = c(tab[2], tab[1]))
## p-value < 2.2e-16
## alternative hypothesis: true odds ratio is greater than 1
## 95 percent confidence interval:
## 857.3396 Inf
## sample estimates:
## odds ratio
## 3358.251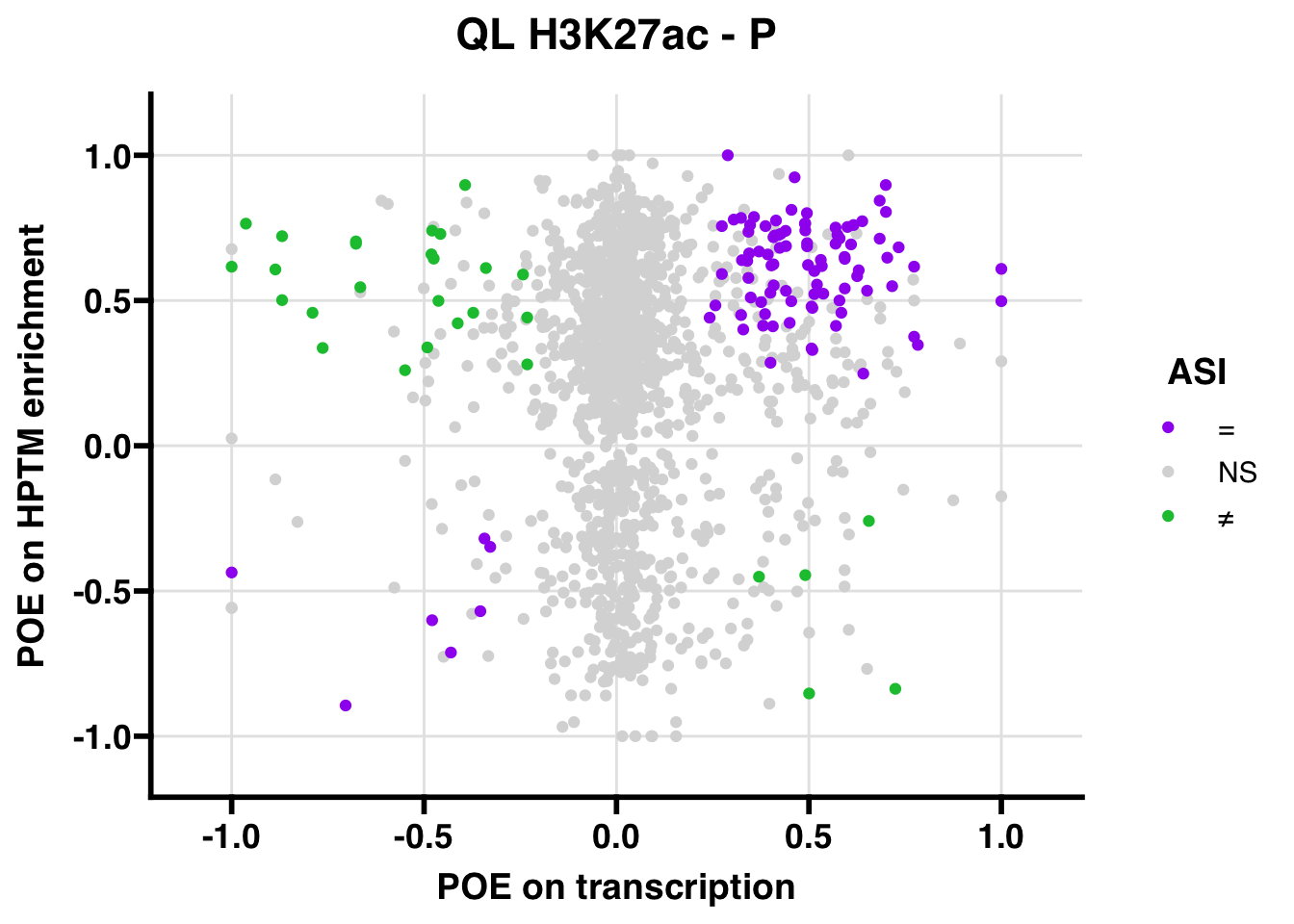
H3K4me3
Gene body
WL
Gene:Peak (categories of allelic bias)
##
## Lineage B:mat Lineage B:NA Lineage B:pat mat:Lineage A mat:Lineage B
## 2 7 3 5 1
## mat:mat mat:NA mat:pat NA:Lineage A NA:Lineage B
## 15 50 21 304 360
## NA:mat NA:NA NA:pat pat:Lineage A pat:Lineage B
## 1432 4824 2141 3 8
## pat:mat pat:NA pat:pat
## 37 84 33Summary of overlaps (00 = neither peak nor gene biased, 01 = gene unbiased, peak biased, 10 = gene biased, peak unbiased, 11 = both gene and peak show allelic bias)
##
## 00 01 10 11
## 8806 27 127 106Fishers exact test of enrichment (do genes showing allelic bias overlap with peaks showing allelic bias)?
##
## Fisher's Exact Test for Count Data
##
## data: data.frame(x = c(tab[4], tab[3]), y = c(tab[2], tab[1]))
## p-value < 2.2e-16
## alternative hypothesis: true odds ratio is greater than 1
## 95 percent confidence interval:
## 180.7667 Inf
## sample estimates:
## odds ratio
## 268.7595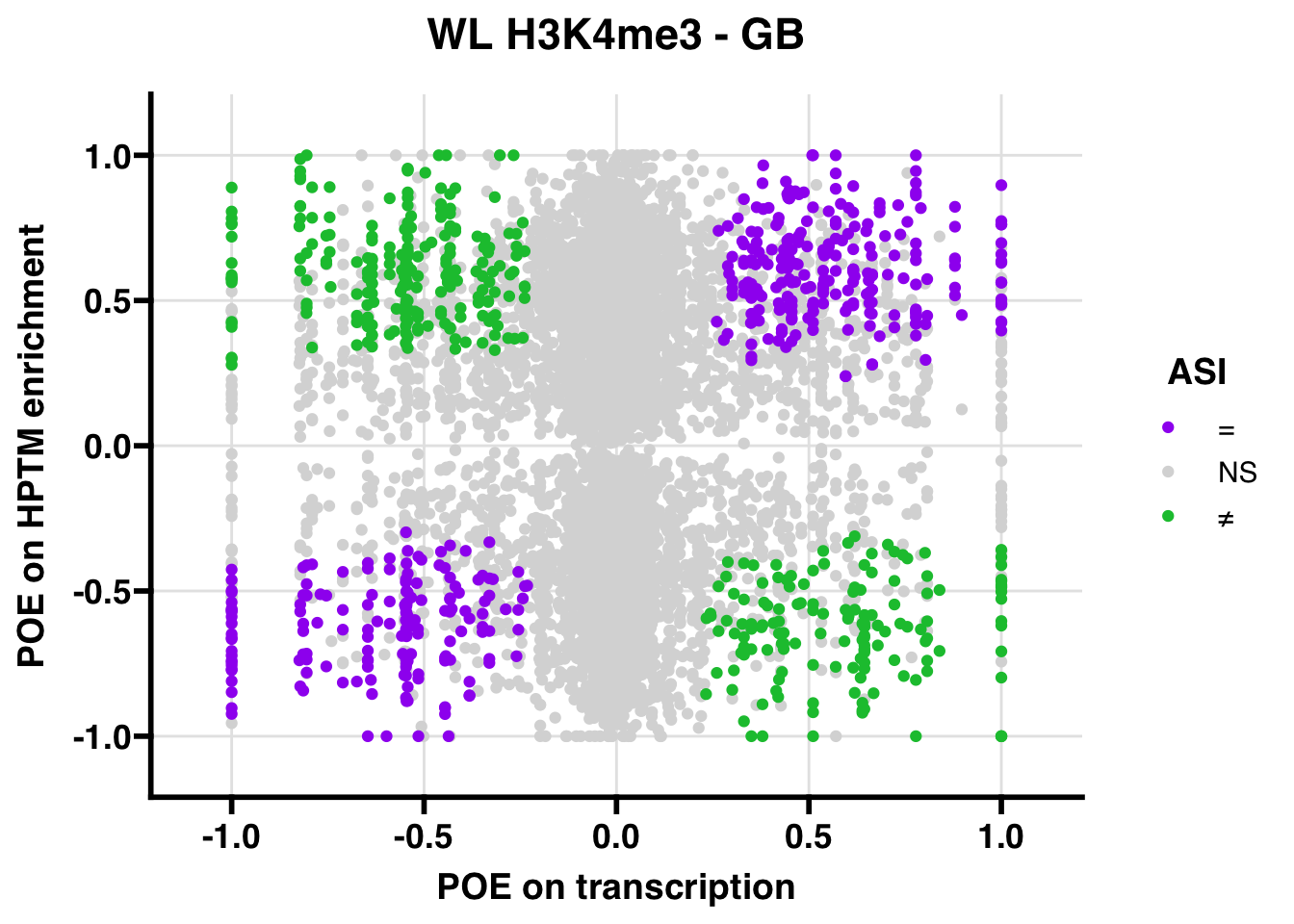
QL
Gene:Peak (categories of allelic bias)
##
## Lineage B:Lineage A Lineage B:Lineage B Lineage B:mat Lineage B:NA
## 1 3 4 18
## Lineage B:pat mat:Lineage B mat:NA mat:pat
## 5 1 22 19
## NA:Lineage A NA:Lineage B NA:mat NA:NA
## 382 422 1003 5775
## NA:pat pat:Lineage A pat:Lineage B pat:mat
## 3836 3 4 5
## pat:NA pat:pat
## 79 89Summary of overlaps (00 = neither peak nor gene biased, 01 = gene unbiased, peak biased, 10 = gene biased, peak unbiased, 11 = both gene and peak show allelic bias)
##
## 00 01 10 11
## 11206 25 81 113Fishers exact test of enrichment (do genes showing allelic bias overlap with peaks showing allelic bias)?
##
## Fisher's Exact Test for Count Data
##
## data: data.frame(x = c(tab[4], tab[3]), y = c(tab[2], tab[1]))
## p-value < 2.2e-16
## alternative hypothesis: true odds ratio is greater than 1
## 95 percent confidence interval:
## 405.6765 Inf
## sample estimates:
## odds ratio
## 609.4468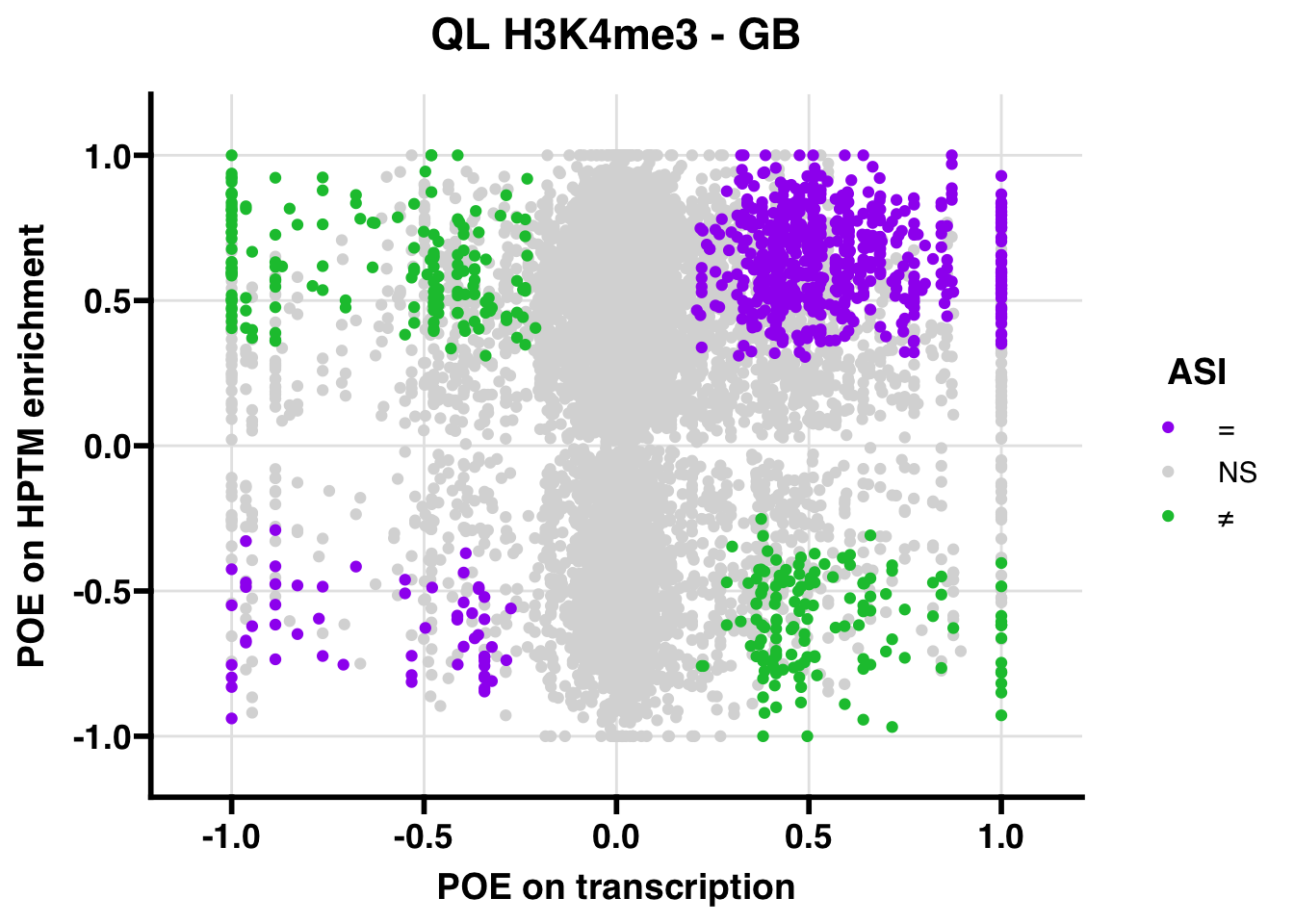
Promoter
WL
Gene:Peak (categories of allelic bias)
##
## Lineage B:mat mat:mat mat:NA mat:pat NA:Lineage A
## 1 1 5 3 73
## NA:Lineage B NA:mat NA:NA NA:pat pat:Lineage A
## 50 247 781 363 1
## pat:mat pat:NA pat:pat
## 4 6 10Summary of overlaps (00 = neither peak nor gene biased, 01 = gene unbiased, peak biased, 10 = gene biased, peak unbiased, 11 = both gene and peak show allelic bias)
##
## 00 01 10 11
## 1490 7 1 18Fishers exact test of enrichment (do genes showing allelic bias overlap with peaks showing allelic bias)?
##
## Fisher's Exact Test for Count Data
##
## data: data.frame(x = c(tab[4], tab[3]), y = c(tab[2], tab[1]))
## p-value < 2.2e-16
## alternative hypothesis: true odds ratio is greater than 1
## 95 percent confidence interval:
## 538.9474 Inf
## sample estimates:
## odds ratio
## 3621.27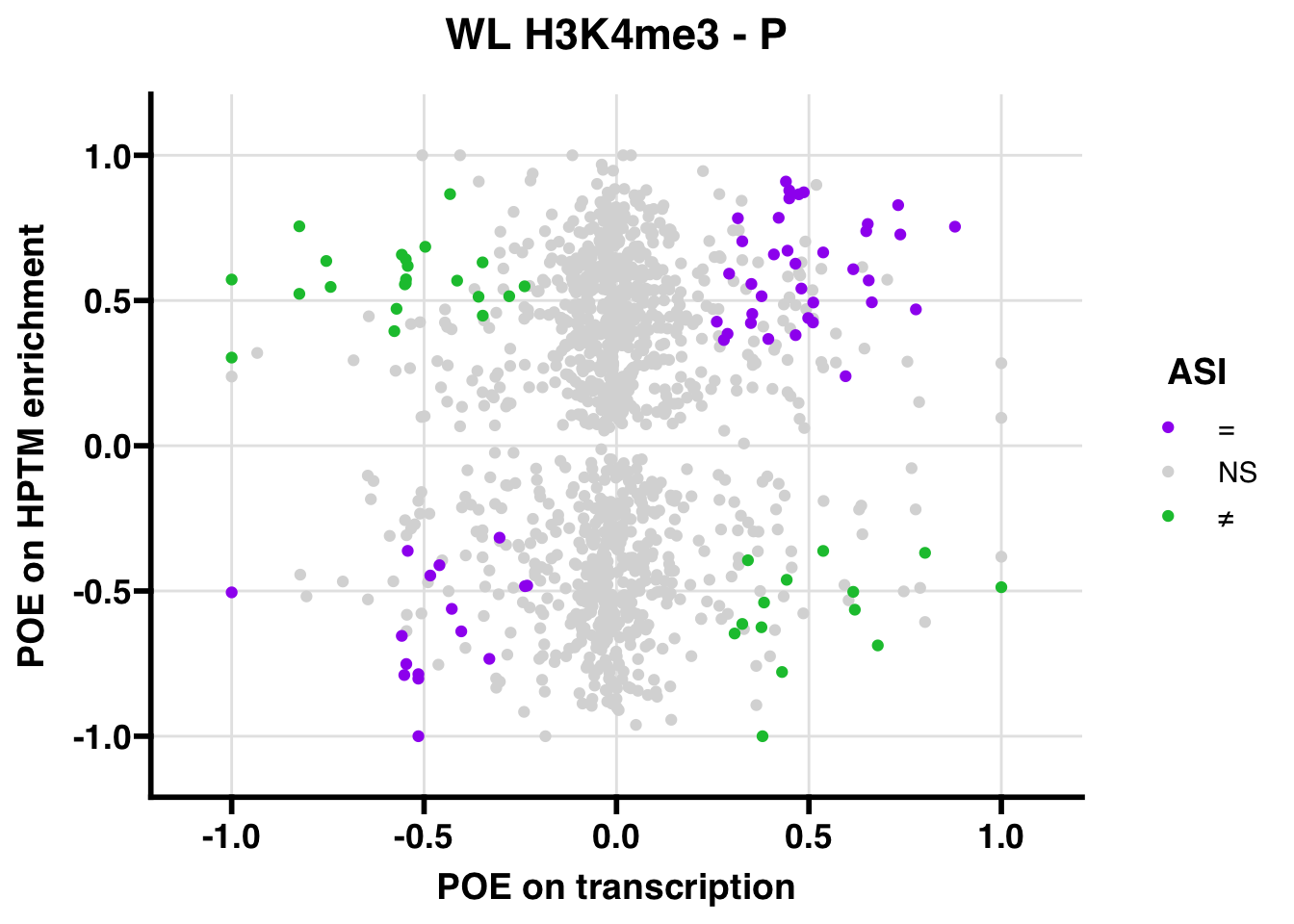
QL
Gene:Peak (categories of allelic bias)
##
## Lineage B:NA mat:NA mat:pat NA:Lineage A NA:Lineage B
## 2 2 6 66 69
## NA:mat NA:NA NA:pat pat:Lineage A pat:mat
## 132 925 645 1 4
## pat:NA pat:pat
## 15 17Summary of overlaps (00 = neither peak nor gene biased, 01 = gene unbiased, peak biased, 10 = gene biased, peak unbiased, 11 = both gene and peak show allelic bias)
##
## 00 01 10 11
## 1801 9 5 27Fishers exact test of enrichment (do genes showing allelic bias overlap with peaks showing allelic bias)?
##
## Fisher's Exact Test for Count Data
##
## data: data.frame(x = c(tab[4], tab[3]), y = c(tab[2], tab[1]))
## p-value < 2.2e-16
## alternative hypothesis: true odds ratio is greater than 1
## 95 percent confidence interval:
## 354.8727 Inf
## sample estimates:
## odds ratio
## 1042.272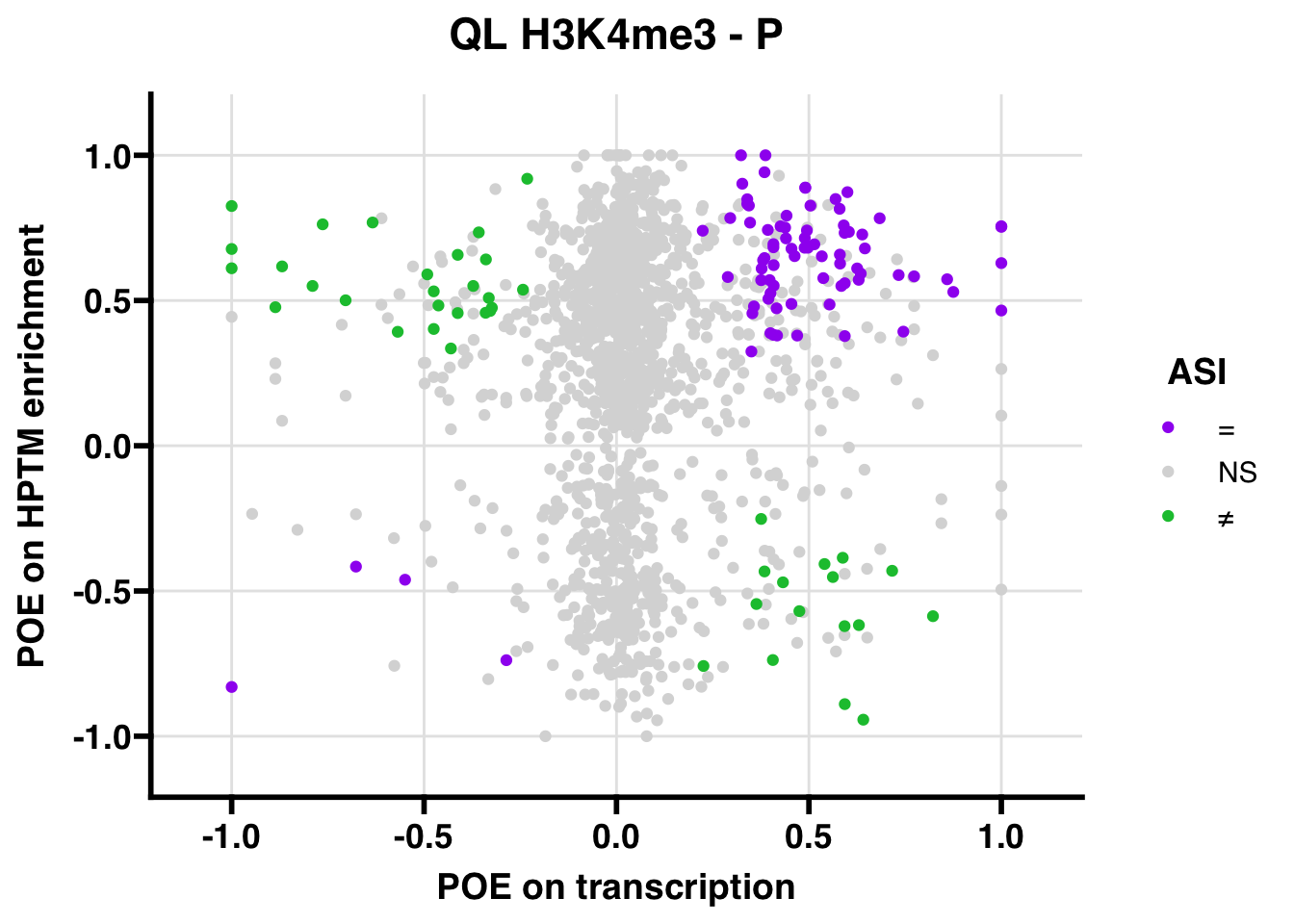
7.3 Logistic regression
H3K27me3 (gene body)
##
## Call:
## glm(formula = OVERLAP.y ~ OVERLAP.x + Group, family = "binomial",
## data = comp.df.sub, na.action = na.exclude)
##
## Coefficients:
## Estimate Std. Error z value Pr(>|z|)
## (Intercept) -5.82864 0.10454 -55.756 <2e-16 ***
## OVERLAP.x 5.70436 0.10300 55.381 <2e-16 ***
## Groupb1W -0.24667 0.10946 -2.253 0.0242 *
## Groupb2Q -0.04658 0.13386 -0.348 0.7279
## Groupb2W -0.26907 0.13136 -2.048 0.0405 *
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## (Dispersion parameter for binomial family taken to be 1)
##
## Null deviance: 8323.7 on 50920 degrees of freedom
## Residual deviance: 3910.9 on 50916 degrees of freedom
## AIC: 3920.9
##
## Number of Fisher Scoring iterations: 8## OR 2.5 % 97.5 %
## (Intercept) 2.942083e-03 2.385536e-03 3.594559e-03
## OVERLAP.x 3.001724e+02 2.461635e+02 3.687337e+02
## Groupb1W 7.813985e-01 6.299726e-01 9.677082e-01
## Groupb2Q 9.544874e-01 7.327419e-01 1.238843e+00
## Groupb2W 7.640882e-01 5.893171e-01 9.865526e-01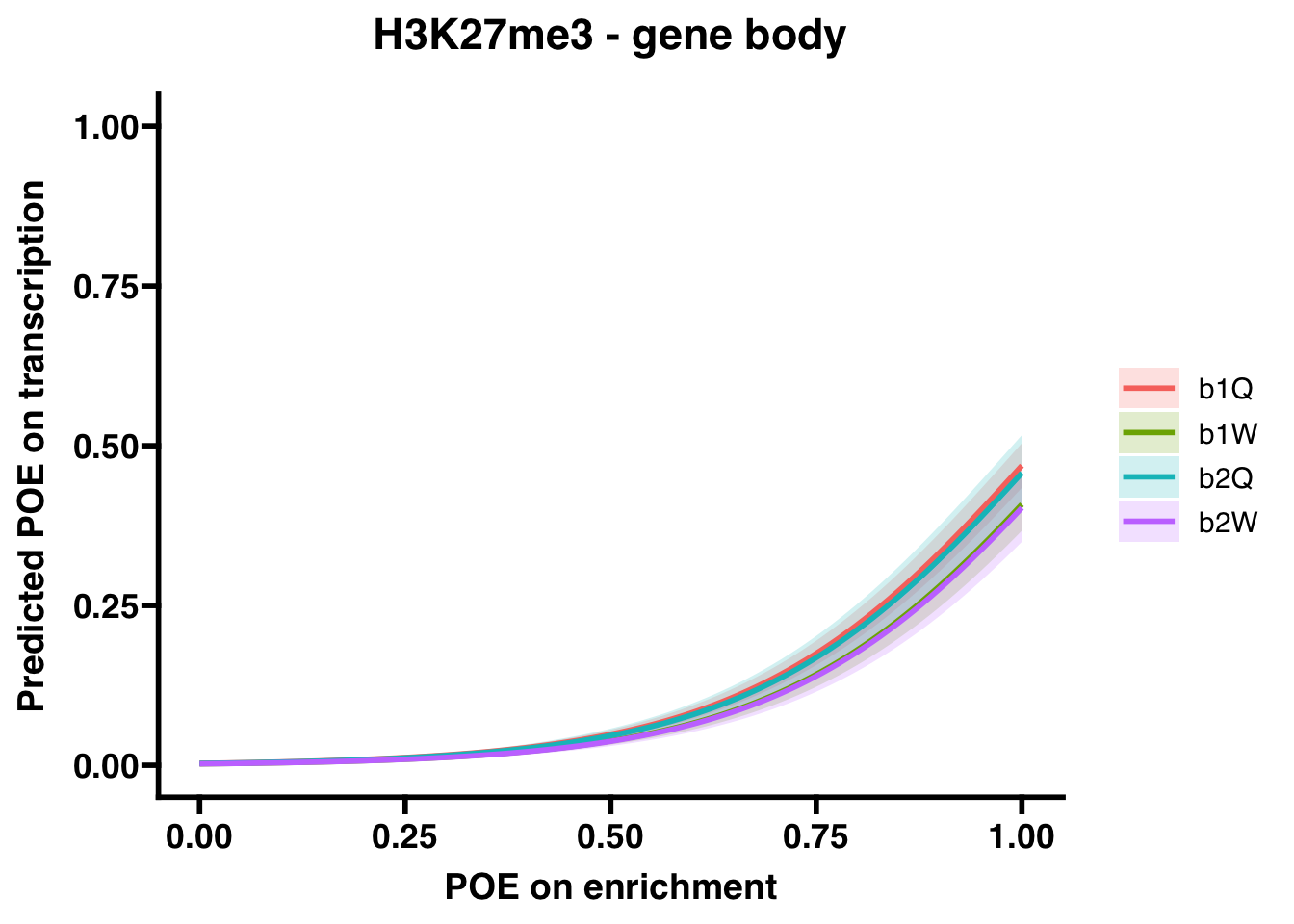
H3K27me3 (promoter)
##
## Call:
## glm(formula = OVERLAP.y ~ OVERLAP.x + Group, family = "binomial",
## data = comp.df.sub, na.action = na.exclude)
##
## Coefficients:
## Estimate Std. Error z value Pr(>|z|)
## (Intercept) -5.75549 0.31152 -18.476 <2e-16 ***
## OVERLAP.x 7.52828 0.34261 21.973 <2e-16 ***
## Groupb1W -0.26110 0.41729 -0.626 0.532
## Groupb2Q -0.53716 0.45621 -1.177 0.239
## Groupb2W -0.06581 0.52996 -0.124 0.901
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## (Dispersion parameter for binomial family taken to be 1)
##
## Null deviance: 1233.94 on 7353 degrees of freedom
## Residual deviance: 374.64 on 7349 degrees of freedom
## AIC: 384.64
##
## Number of Fisher Scoring iterations: 8## OR 2.5 % 97.5 %
## (Intercept) 3.165344e-03 0.00163964 5.583912e-03
## OVERLAP.x 1.859898e+03 979.24688378 3.769529e+03
## Groupb1W 7.702022e-01 0.33622761 1.742170e+00
## Groupb2Q 5.844072e-01 0.23538931 1.418525e+00
## Groupb2W 9.363044e-01 0.31627339 2.563678e+00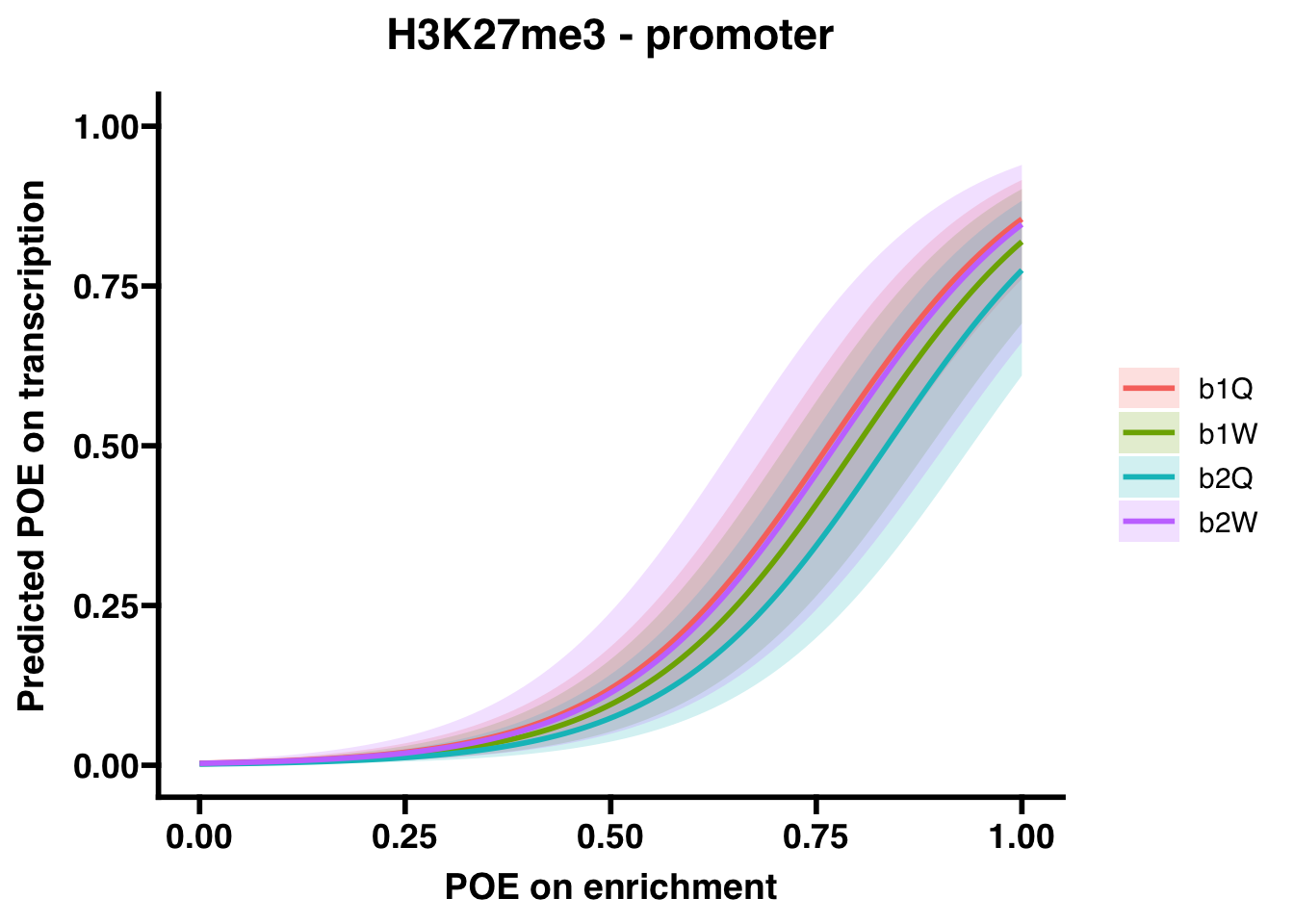
H3K27ac (gene body)
##
## Call:
## glm(formula = OVERLAP.y ~ OVERLAP.x + Group, family = "binomial",
## data = comp.df.sub, na.action = na.exclude)
##
## Coefficients:
## Estimate Std. Error z value Pr(>|z|)
## (Intercept) -5.68839 0.09942 -57.217 <2e-16 ***
## OVERLAP.x 5.62249 0.09830 57.197 <2e-16 ***
## Groupb1W -0.20618 0.10566 -1.951 0.051 .
## Groupb2Q 0.08954 0.12566 0.713 0.476
## Groupb2W -0.12049 0.15052 -0.801 0.423
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## (Dispersion parameter for binomial family taken to be 1)
##
## Null deviance: 8533.4 on 47753 degrees of freedom
## Residual deviance: 4071.8 on 47749 degrees of freedom
## AIC: 4081.8
##
## Number of Fisher Scoring iterations: 8## OR 2.5 % 97.5 %
## (Intercept) 3.385034e-03 2.773866e-03 4.096361e-03
## OVERLAP.x 2.765771e+02 2.287886e+02 3.364209e+02
## Groupb1W 8.136896e-01 6.610398e-01 1.000369e+00
## Groupb2Q 1.093668e+00 8.538520e-01 1.397763e+00
## Groupb2W 8.864828e-01 6.578226e-01 1.187315e+00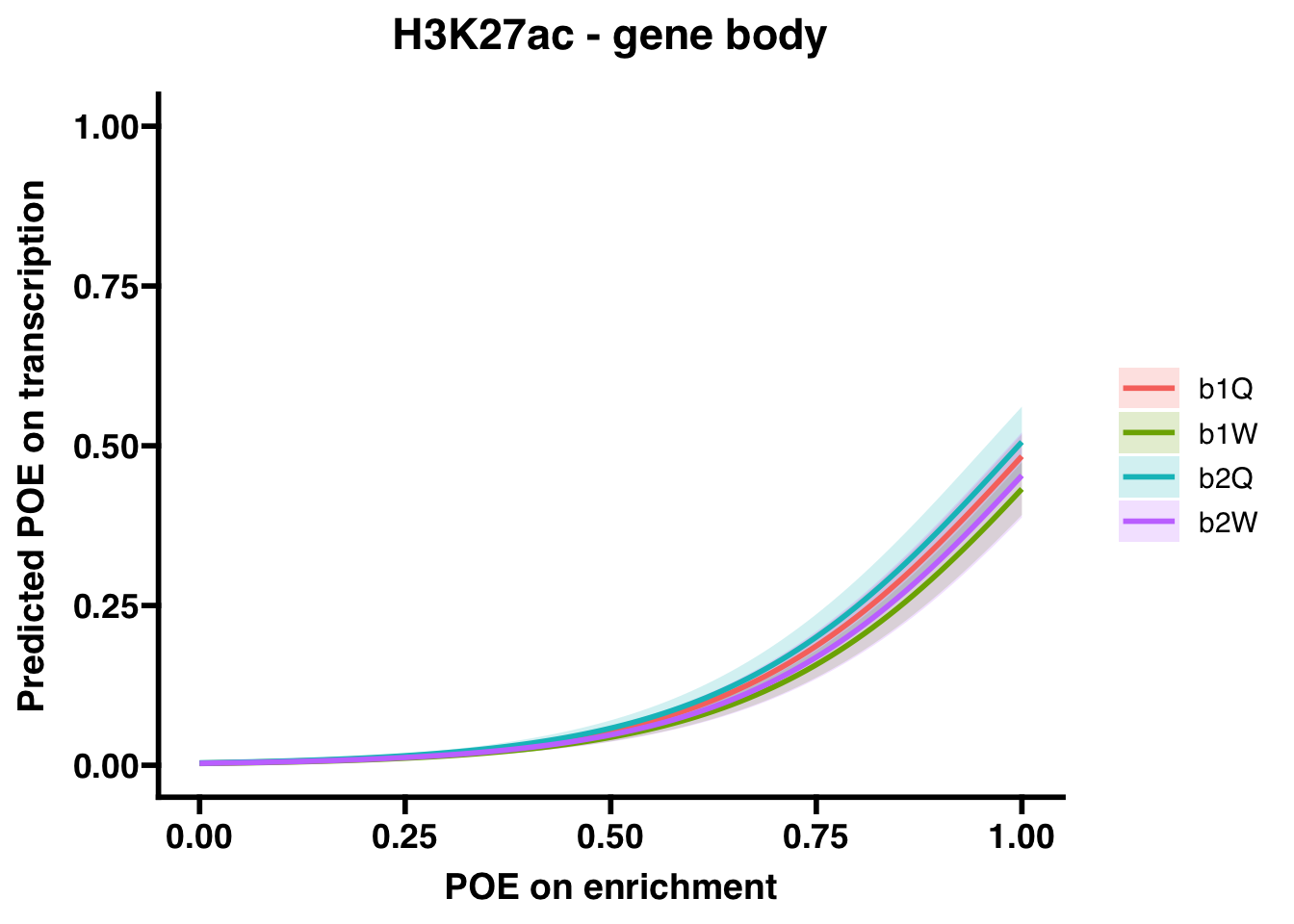
H3K27ac (promoter)
##
## Call:
## glm(formula = OVERLAP.y ~ OVERLAP.x + Group, family = "binomial",
## data = comp.df.sub, na.action = na.exclude)
##
## Coefficients:
## Estimate Std. Error z value Pr(>|z|)
## (Intercept) -5.63126 0.28548 -19.726 <2e-16 ***
## OVERLAP.x 7.12448 0.29693 23.994 <2e-16 ***
## Groupb1W -0.04749 0.36865 -0.129 0.898
## Groupb2Q 0.01817 0.39244 0.046 0.963
## Groupb2W -0.01675 0.48733 -0.034 0.973
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## (Dispersion parameter for binomial family taken to be 1)
##
## Null deviance: 1360.28 on 7500 degrees of freedom
## Residual deviance: 474.87 on 7496 degrees of freedom
## AIC: 484.87
##
## Number of Fisher Scoring iterations: 8## OR 2.5 % 97.5 %
## (Intercept) 3.584039e-03 1.975511e-03 6.060581e-03
## OVERLAP.x 1.242006e+03 7.086525e+02 2.275533e+03
## Groupb1W 9.536245e-01 4.608848e-01 1.968569e+00
## Groupb2Q 1.018332e+00 4.687536e-01 2.197517e+00
## Groupb2W 9.833923e-01 3.694404e-01 2.502751e+00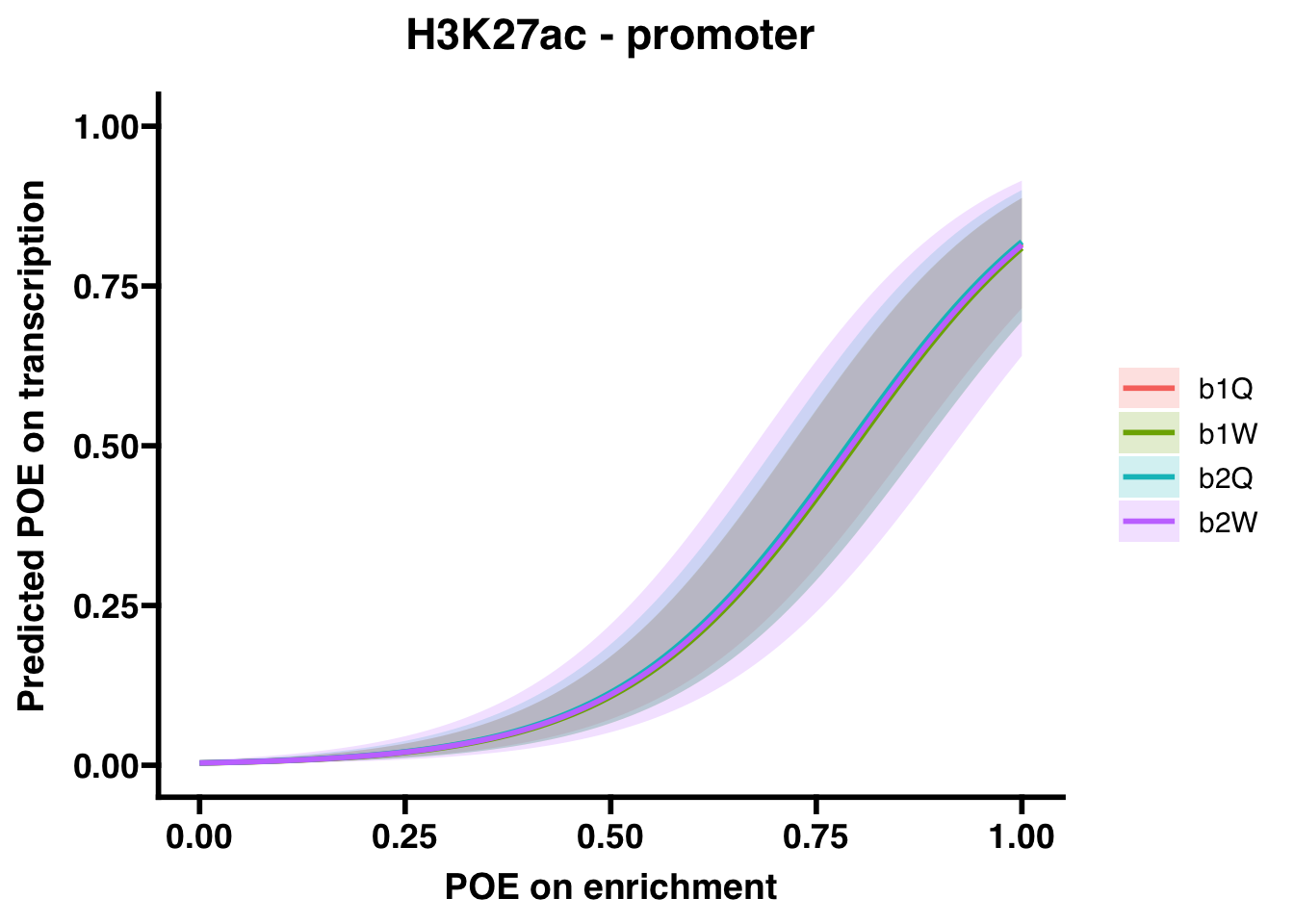
H3K4me3 (gene body)
##
## Call:
## glm(formula = OVERLAP.y ~ OVERLAP.x + Group, family = "binomial",
## data = comp.df.sub, na.action = na.exclude)
##
## Coefficients:
## Estimate Std. Error z value Pr(>|z|)
## (Intercept) -6.089231 0.118185 -51.523 <2e-16 ***
## OVERLAP.x 6.041222 0.112453 53.722 <2e-16 ***
## Groupb1W -0.209966 0.117669 -1.784 0.0744 .
## Groupb2Q 0.233025 0.135792 1.716 0.0862 .
## Groupb2W -0.008768 0.134921 -0.065 0.9482
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## (Dispersion parameter for binomial family taken to be 1)
##
## Null deviance: 8136.4 on 48209 degrees of freedom
## Residual deviance: 3495.8 on 48205 degrees of freedom
## AIC: 3505.8
##
## Number of Fisher Scoring iterations: 8## OR 2.5 % 97.5 %
## (Intercept) 2.267151e-03 1.788273e-03 2.842929e-03
## OVERLAP.x 4.204063e+02 3.387469e+02 5.266395e+02
## Groupb1W 8.106115e-01 6.431508e-01 1.020280e+00
## Groupb2Q 1.262412e+00 9.667989e-01 1.646939e+00
## Groupb2W 9.912702e-01 7.600011e-01 1.290169e+00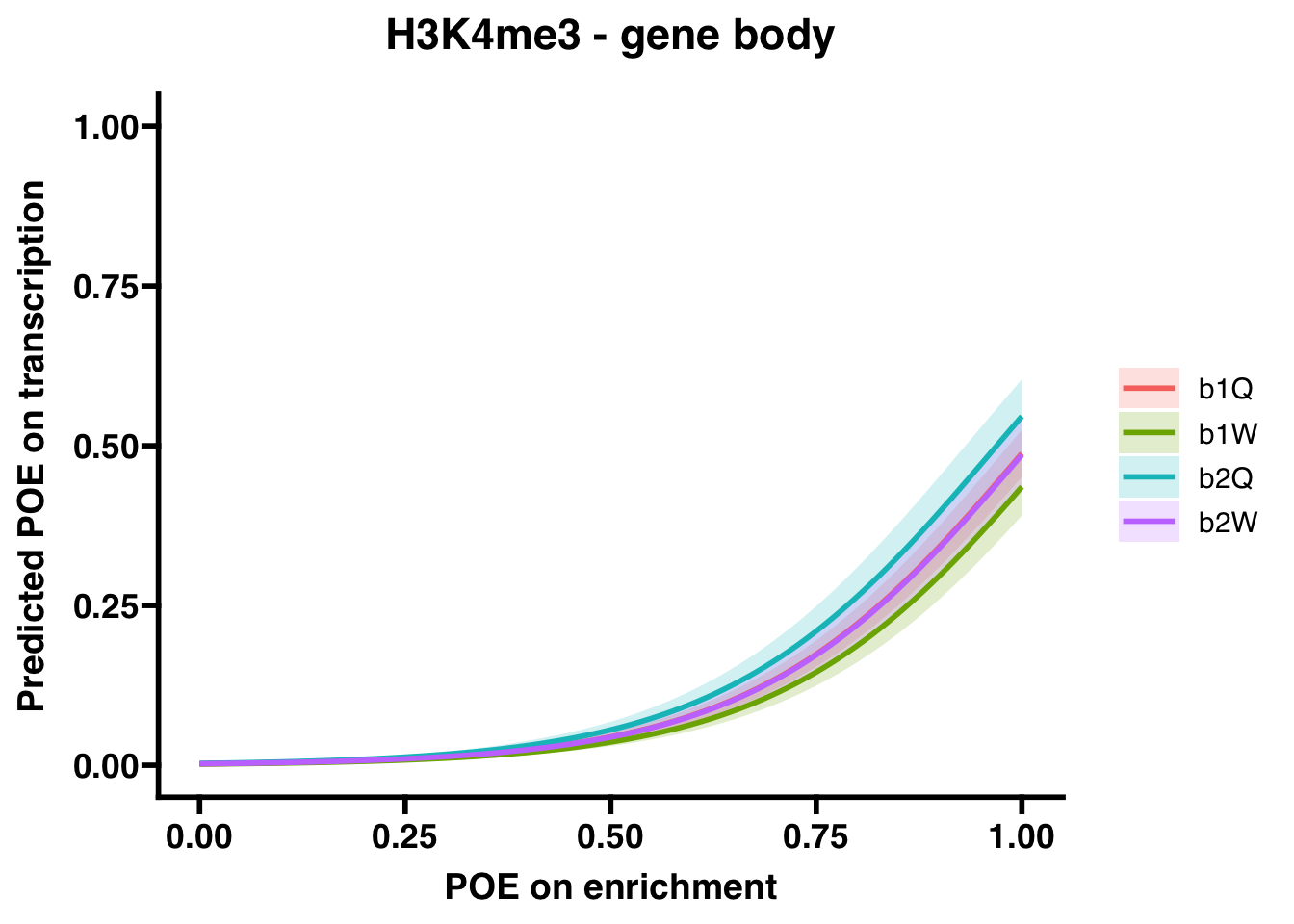
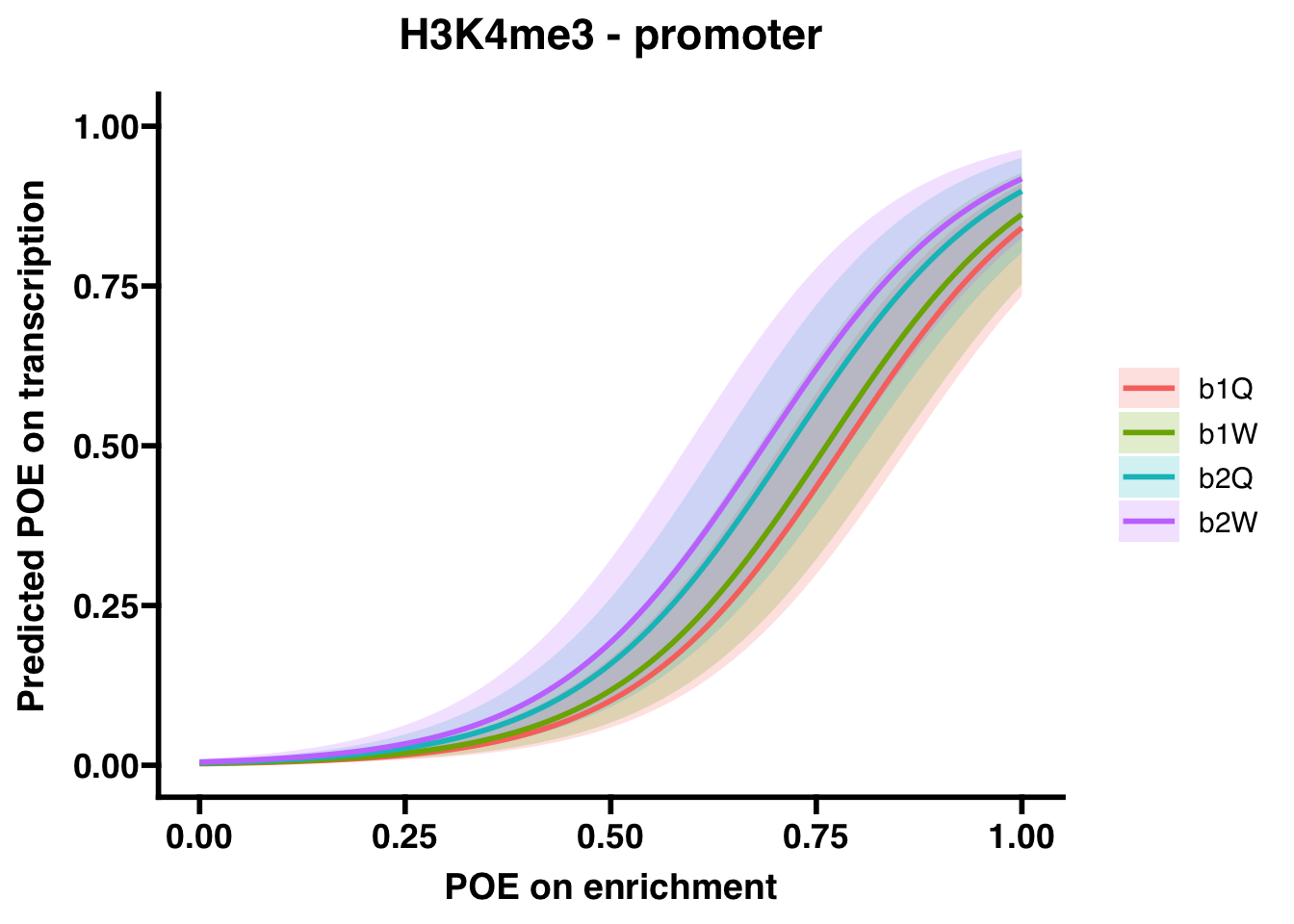
H3K4me3 (promoter)
##
## Call:
## glm(formula = OVERLAP.y ~ OVERLAP.x + Group, family = "binomial",
## data = comp.df.sub, na.action = na.exclude)
##
## Coefficients:
## Estimate Std. Error z value Pr(>|z|)
## (Intercept) -6.0322 0.3567 -16.910 <2e-16 ***
## OVERLAP.x 7.6968 0.3422 22.495 <2e-16 ***
## Groupb1W 0.1652 0.4352 0.380 0.704
## Groupb2Q 0.5159 0.4434 1.164 0.245
## Groupb2W 0.7490 0.4693 1.596 0.110
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## (Dispersion parameter for binomial family taken to be 1)
##
## Null deviance: 1433.79 on 7359 degrees of freedom
## Residual deviance: 437.22 on 7355 degrees of freedom
## AIC: 447.22
##
## Number of Fisher Scoring iterations: 8## OR 2.5 % 97.5 %
## (Intercept) 2.400151e-03 1.134377e-03 4.603934e-03
## OVERLAP.x 2.201259e+03 1.163417e+03 4.474247e+03
## Groupb1W 1.179619e+00 5.026020e-01 2.796025e+00
## Groupb2Q 1.675221e+00 7.069031e-01 4.063030e+00
## Groupb2W 2.114952e+00 8.439543e-01 5.373705e+00